1) There is no change to Schrodinger license and usage from Eric's prior document. Currently the Desktop and several linux nodes have been properly set up to run Schrodinger.
2) Other than those files in the Dropbox, all the data/results on the Desktop is under C:\Users\plin\Documents. All the raw Schrodinger data/analysis is in the project directories under Documents\Schrodinger\Ping. Most of PCR related codes/reference can be found under Documents\PCR_Software. MD analysis done using Desktop is located under Documents\MD_works and Documents\Work. Contents of all other directories can be easily identified by the name.
All the raw data/analysis using linux nodes can be found under pmcat-gpu1:/home2/plin/work. Codes for MM-PB(GB)SA calculation using dock poses in pdb format can be found under /home2/plin/bin. Other than the backup files and LEA3D test runs, MD simulations, Schrodinger jobs (on linux), and data analysis results are under /home2/plin/work/project01. The name of directory usually tells what is content is about.
3) Passwords have been sent to Wei for accessing the Desktop/linux nodes.
RC (06/11):
The points below should ideally be addressed prior to Tues 6/16. If any issues are foreseen or clarification needed, please indicate in advance.
--Questions on subtask b):
In an extension of this report, also provide the following in order to analyze effect of loop conformation on the ternary complex: prepare the structure of the ternary complex (NAD+ before rxn) starting from 4FVT with loop replacement from 4BVG, carry out sidechain optimization including loop residues (compute RMSDs of these sidechains with respect to those from 4BVG, and compare to sidechain validation study for 4BVG), provide single point score. PL: The calculations here include 4FVT, 4FVT w/ loop substitution, w/ or w/o sidechain predictions. Do we need to explore single or short-chain sidechain prediction here?
RC: In the first draft of the report do multi-sidechain prediction on the complete residue set for the substituted loop and environment (e.g., with MC prediction as you did in the earlier validation study reports). Indicate whether you observe any of the same types of interactions that were associated with energy errors identified in the validation studies (see below regarding tabulation of those energy errors). Report the RMSDs of side chains including those in loop environment with respect to the native 4BVG structure and comment on differences.
Note that these calculations must include NAD+ ligand. The required calculations involve the use of SIRT3/NAD+ from 4FVT with loop replacement on residue 155-178.
(I noticed a document called "Sidechain prediction for SIRT3/INT/NAM converted from 4FVT with sidechain replacement on residue 155-178 from 4BVG" in dropbox.)
In case of high energy, substitute C pocket sidechains w Ala or reduce vdW radii while minimizing NAD+, then carry out sidechain optimization (as in C pocket ligand structure preparation task), followed by short MD sampling if needed. PL: Carry out extra modification before sidechain prediction, right? Is MD desirable here?
RC: Yes the extra modifications (if required) are to be carried out before sidechain prediction. Report the single point results prior to running MD. MD can run while you are out of town. (Please note that the C pocket ligand structure preparation task was not yet completed.)
Compare single point energy to that of minimized 4FVT in this report (and then also compare MM-GB(PB)SA energies with MD sampling as part of MD task below). If energy of 4BVG loop conformation is higher than that of 4FVT loop conformation, identify interactions that are responsible for the difference with respect to the energy gap in the INT:NAM complex, (PL: why SIRT3:INT:NAM complex?)
RC: For the SIRT3:INT:NAM complex (after reaction), the 4BVG loop conformation appears energetically favored (energy gap between 4BVG and 4FVT loop conformations was estimated by MD), whereas for the SIRT3:peptide:NAD+ complex (before reaction), the 4FVT loop conformation appears to be preferred. We seek to determine whether the computations provide a consistent sign for the energy gap on the SIRT3:peptide:NAD+ complex and identify the interactions responsible for the differences in the energy gaps for the two complexes.
Comparison can be made based on the existing results for SIRT3:INT:NAM and ultimately updated after the task “After updating MD and sidechain opt short reports and discussion with RC, restart SIRT3:INT:NAM MD simulations starting from the structures based on latest sidechain opt and NAM complex preparation results”
and cross-reference the nicotinamide contacts report if relevant. Comment on possible role of carba-NAD if relevant.
--Report on sidechain prediction for 4BVG with loop replacement from 4FVT:
Edits to this report are needed, as summarized below:
- The sidechains are being optimized in a non-native environment. Hence it is important to compare the sidechain conformations of the environment to those from a native structure. In the xls, you have reported RMSDs for the sidechains in the environment (e.g., residue 231), which were not substituted; are these RMSDs with respect to 4FVT after alignment or with respect to 4BVG? The former should also be reported. Special attention should be paid to those differences that originate due to interactions with the nonnative environment (e.g. sidechains in the loop environment that adopt conformations different from those in 4FVT), which should be highlighted.
To accompany the xls already provided, please comment on the comparison of the conformations of sidechains in the loop environment to those from 4BVG after optimization in the sidechain prediction validation study on native 4BVG (validation set 2 below). For example, if the RMSDs in this xls are with respect to native 4BVG, it may be more informative to indicate the RMSDs with respect to sidechain-optimized 4BVG, since some of the discrepancies with respect to native may be the same in both studies.
If there are any of the same types of interactions that were associated with energy errors identified in the validation studies, they should be indicated.
(This study might be best done for 4FVT with loop replacement from 4BVG, since 4BVG is the native structure without the NAD+ ligand, and we are not including NAD+ in these studies; but this is not necessary for now.)
The above can be done after completion of subtask b), since the analysis required is very similar. In case of time constraints, complete subtask b) and the edits requested to sidechain predictions validation set 2 (below) first.
--Report on sidechain prediction validation set 2
RC: Edits to this report are needed, as summarized below:
- The xls is useful for the complete dataset, but we need to organize the results in terms of:
a) sampling errors (total energy higher than native for incorrect structure predictions) where energy errors for sidechain subsets have not been identified
b) energy errors (total energy lower than native for incorrect structure predictions) where sampling errors for sidechain subsets have not been identified
c) overall sampling errors that also contain contributions from energy errors that have been identified as part of the small-set sidechain prediction (if any). For example, in multi-sidechain prediction of the whole loop with the default algorithm, the total energy may be higher than the native, but there may still be energy errors for subsets of sidechains that lower the energy for the incorrectly predicted structures. By manually correcting these energy errors, we could isolate the energy gap due to the sampling errors (latter optional).
d) overall energy errors that also contain sampling errors that have been identified as part of the small-set sidechain prediction (if any).
Please aim to fit each of the prediction data should fit into one of the above categories. A table can be used if convenient.
-Color coding (green/yellow) in the xls should be explained
- Figures are unclear; it is necessary to present them in a form that might ultimately be suitable for publication. The relevant interactions – which are different between native and predicted structures – should be magnified and presented in more than one subfigure if necessary for clarity. The above will help us better analyze the types of energy errors observed. Also, in some cases, an interacting residue pair is mentioned but only one member of the pair is shown.
As noted in single sidechain prediction comments, a table can be used to organize the energy errors and list the types of errors observed in each case
- Some comments in the reports state that sampling was insufficient for multi-sidechain prediction, but do not specify what algorithm was used in those cases. I assume the default algorithm was used in those cases.
PL(06/09): Sidechain prediction validation results for 4BVG w/ loop substitution was summarized under Dropbox\PMC-AT PLIN\
Sidechain prediction for 4BVG with sidechain replacement on residue 155-178.docx and
4BVG-s155-178-sp-set1_DATA.xlsx .
PL(06/05): The estimate time for sub tasks were estimated. Some depends on whether to run certain calculation or not, and can change significantly.
Upload sidechain prediction-validation_set2.docx to Dropbox, and the 4BVG-sp-set2_DATA.xlsx, which cover the observation on short chain sidechain prediction results.
RC (6/4): The schedule for the three tasks that were divided into subtasks has not been updated with estimated times for each subtask. This was to be completed by 6/2.
See below - the estimated time for each subtask is to be listed in the same cell to the right of the subtask.
I will need to review these before approval.
PL(6/2): scheduled updated. Under Dropbox\PMC-AT PLIN\, Single sidechain prediction results on 4BVG.docx was updated; 4BVG-ssp-DATA.xlsx was added.
Currently working on short chain (3-5 residues) sidechain prediction.
Residues in contact with NAM is updated including the referenced papers. (see
Residues in contact with NAM_updated.docx).
RC (6/1): Updated schedule with times required for each specified subtask -- already indicated previously below - is required as a first priority.
A posting to the wiki indicating that the updates have been uploaded should be made by Tues.
RC (5/28):
Tasks have been divided into subtasks that should be updated incrementally in the same reports as they become available.
The estimated times required for these subtasks should be provided separately in the task list (beside the subtask, in the same cell).
The estimated time required for some of the tasks appears too long in some cases. A total of 5-6 weeks full-time work
has been estimated for 3 tasks. One of these is based entirely on existing simulation data.
Single side chain prediction report has been updated with comments for next steps for side chain prediction validation task.
Side chain algorithm options should be listed as discussed.
All previously posted comments (including those in original side chain prediction report) should be answered/addressed.
Wiki should indicate when dropbox has been updated with reports.
RC (5/26):
-Task list updated on dropbox. Estimated times for tasks need to be added (PL).
Time for meeting this week will be determined based on this schedule.
-By end of next week, we should be able to complete up to but not necessarily including all of the following task:
"After updating MD and side chain opt short reports and discussion with RC, restart SIRT3:INT:NAM MD simulations starting from the structures based on latest side chain opt and NAM complex preparation results"
Please specify schedule accordingly.
-For purposes of C pocket ligand structure preparation task, the following needs some attention. See "Mechanism of Sirtuin Inhibition by Nicotinamide: Altering the NAD+ cosubstrate specificity of a Sir2 Enzyme" (Wolberger, uploaded to wiki previously). Review Wolberger NAD/NAM contacts for Sir2's and see esp pg. 859 for differences in binding modes of NAM and nicotinamide moiety in NAD+ (given that the latter is used for alignment in preparation of some of our NAM structures). In our earlier discussions, it was indicated that there are no NAM bound structures? Please clarify vis-a-vis the xtal structures discussed in this paper. Do these xtal structures have any implications for the effect of the loop conformational change between ternary and intermediate structures on the contacts made by NAD (ternary complex) vs NAM (intermediate complex)? These points may later be relevant for NAM binding affinity calculations. PL's earlier report on NAM contacts from 1st section of task list (below) should be updated, mentioning these points in the context of the differences between NAD and NAM nicotinamide binding modes, to accompany the upcoming C pocket ligand structure preparation short report. In the NAM contacts report (and elsewhere whenever possible), make revisions to figures like those for residues in contact w nam, providing a representation displaying h bonds or table columns indicating the type of contact for each residue (this can be checked against XG's annotated sequence alignment, which specifies the roles of many important residues in terms of their catalytic or binding role).
XG(5-27) Sequence alignment_02.09.2015.pdf
Also, we previously concluded that the conjecture about strain in NAD was not supported by our computations (via MM calculations for various conformations) - we should link to that analysis on the wiki and indicate if it was for Sir2 or SIRT3.
-If time permits next week and you are waiting for additional input from RC on the above task on restarting SIRT3:INT:NAM MD simulations, move on with task list section 1 loop scoring analysis (energy and sampling errors, with analysis of sources of energy errors), based on similar principles to side chain opt, as well as possible helical sampling for Sir2Tm ternary loop (the task list part 1 has been updated to specify the latter).
PL(05/22): Files uploaded to Dropbox on sidechain validation studies.
RC (5/26):
Comments on sidechain validation studies including immediate priorities for this week required for further discussion/meeting have been updated in
the doc file w suffix PL_RC.
These tasks are priorities for this week; need to know when they will be finished to stay on schedule for incorporation into paper draft – specify in tasks list under the side chain validation task. Meet mid week if possible after another update to this report.
Some selected comments from the annotated document:
-No side chain energy error analysis? Analysis of energy errors for smaller number of residues as indicated, including single sidechain prediction (ssp)?
-Would like to see ssp on each of the yellow residues identified, along with analysis of types of energy errors
-Specify the energy gap between native and predicted structures for ssp energy errors
-Focus on smaller sets of side chains to reduce sampling errors
-Provide a table listing each of the ssp energy errors and showing the native structure, the predicted structure, and a comment on the type of interaction that was scored incorrectly
-Check for possibility of missed bridging waters, protonation state errors, etc
RC (5/19): Task list is updated on dropbox.
Short report on MD has been reviewed.
Please add estimated time required for updated version of each short report that is indicated on the task list.
RC (5/15):
1) Side chain opt validation (priority)
- No RMSD data was presented in the side chain optimization report thus far. Please see original email, which describes what should be provided and why, and add that before we discuss. Related, there is also no discussion of energy vs sampling errors (this was one of the most important parts of the analysis). This should be added.
- Please enumerate the types of energy errors identified in your side chain opt short report. Wherever you found an energy error (assuming you found some), please reduce the number of side chains subjected to optimization so as to determine whether the energy error remains for a single side chain.
- Can we do more extensive side chain sampling in the validation studies where you identified sampling errors? What are the adjustable parameters for side chain sampling? Please report on these parameters for MC side chain optimization and indicate whether we can access the low energy rotamers sampled by MC optimization (if so, how many; these may later be useful in several contexts, including C pocket ligand complex preparation).
- In the first table under b) in the report, did you subject the same side chains listed at the top of the report to optimization? So, you only have a limited dataset (apparently, consisting of four members - two each for 4FVT and 4BVG).
PL(05/20): I am currently running side chain prediction with various options on both 4FVT (SIRT3/carba-NAD+/ac-LYS complex) and 4BVG (SIRT3/INT complex) using limited set of residues in the optimization. RMSD calculations were performed and will be put together in the report before Friday.
2) C pocket side chain optimization for ligand complex preparation
- The emphasis here is not on loop substitution, but on application of side chain optimization to complex preparation for our planned, higher accuracy NAM binding affinity calculations. As noted, a comparison between sampling methods previously discussed is the priority. Energies obtained by each sampling method should be compared. RMSDs should be provided.
- The focus should hence be on simple case starting from 4BVG without loop substitution. [I believe you included other results you had obtained (e.g., for loop substitution) prior to my email clarification regarding this task. These can be separated from the report for now for purposes of clarity. When we revisit loop substitution, it will have to be analyzed properly in the context of the side chain opt validation results, as noted.]
- The number of side chains subjected to optimization need not be as large as that for the loop substitution studies. Please consider three radii around NAM (small, medium, large) for side chain optimization, because energy and sampling errors can arise for larger radii. Provide the energies and RMSDs for each, and indicate whether you see an increase in energy with increasing radius in any case (indicating a sampling error). If not, there still may be energy errors for larger radii, and we will consider this after receiving the complete side chain validation analysis requested above.
- You should also compare to no side chain optimization, as well as the preparation method applied in the PLOS paper. We want to compare the energies to see if side chain optimization is essential for proper NAM complex preparation.
- The Ala-based sampling method (like IFD) has apparently not yet been run. This might be carried out as an IFD docking job. Since IFD does not use alignment to generate the NAM pose, please report the RMSDs of several of the top poses and associated side chain conformations generated by this method with respect to a representative structure, along with their energies.
- A couple of other sampling methods may also be considered in the next step (these will be discussed after the short reported is updated with the results above).
PL(05/20): The calculations will be scheduled once the validation runs are completed and analyzed. I plan to run SIRT3/INT/NAM using only 4BVG as starting point, and try different protocols for predicting NAM binding and overall energy (IFD-like protocols, or NAM placements first followed by sidechain prediction, or sidechain prediction first followed by NAM placement, or reduced vdw radii in the docking, or Ala substitution before docking, or iterative approach to locate the minimum.)
3) MD/MM-GB(PB)SA
- When presenting results from MD simulations previously run, please include a comparison of the ensemble average structure to the starting structure prepared via loop substitution and side chain optimization
- Please also set up MD simulations on the SIRT3/INT complex (no NAM) prepared from 4FVT - I would like to use this prepared
complex to check whether catalytically important side chains display the appropriate contacts/H-bonding patterns required for the next step of catalysis (recall that these were studied as part of the tasks in the first section of the task list). This is another way to check the validity of our structure preparation methods. If you have any questions about the latter, let me know.
Regarding loop prediction, which we will return to soon, I would like to verify one point in advance lest I forget later: please confirm that our previous studies all included crystal packing. There are several other studies reporting success in loop predictions around 10-12 residues in length.
RC (5/8):
a) C pocket side chain optimization in presence of NAM: the emphasis here is comparison of energies of the method used for NAM complex preparation in the PLOS paper (as I recall docking to 4BVG, which has Phe in C pocket, followed by minimization) and the more recently applied method, wherein we do side chain optimization as well. The latter could be done in several ways. One would be straight docking followed by side chain optimization; another would be mutating the residues to alanine, docking the NAM, and then replacing the residues and doing side chain optimization (as in IFD). As you said, iteration could subsequently be applied as is sometimes done in IFD.
This study is a particularly application of induced fit, which will later (in part 3 of tasks list) be used to estimate binding affinities of C pocket binding ligands.
As we discussed it is an opportunity to study the role of the Phe and Arg in the NAM binding affinity/sensitivity of sirtuins. We will later also be applying mutations in silico to explore these effects.
I believe you have already done straight docking followed by side chain optimization; if you prefer you can write this comparison up and then we can set up the alanine mutation method (no iteration) thereafter.
If you like, you can redo you previous study with a more careful consideration of which side chains to include in optimization given the goals of this study.
b) side chain optimization validation: the purpose of this study is to provide some preliminary data for the analysis of the accuracy of side chain optimization
in our mechanistic (and ultimately design) studies of sirtuins in this paper; energy vs sampling errors provide more insight into the causes of errors.
As a first step, I indicated that we can apply multiple side chain optimization to the 4FVT and 4BVG xtal structures -without NAM placement or loop substitution- and check for such errors.
Because sampling errors are more severe when more side chains are included in a single optimization, I suggested grouping them in subsets, each of which is subjected to multiple side chain optimization. This also provides a larger dataset. Comparison of energies and reporting of RMSDs with respect to native (with minimization) are important. Most likely there will be several more sampling errors identified (side chain optimization energy higher than native, with imp structural differences) but I would like to see if there are any examples of energy errors (side chain optimization energy lower than native, with imp structural differences). If not, we can then proceed to look at smaller subsets of side chains to determine whether energy errors occur. For example, for single side chains, sampling errors are rare. (Having found energy errors, we can subsequently investigate their causes if desired.)
So far, you have identified several sampling errors in nonnative structures, where the energy of the side chain optimized structure was higher than that of the structure without side chain optimization. Use of side chain optimization in native structures may allow the identification of energy errors as well.
This task has implications for the side chain optimization with loop substitution (where backbone degrees of freedom need not be sampled) that you reported in the short reports so far, but loop substitution need not be used in the validation analysis, for the reasons mentioned above. For example, we can include the results of the side chain optimization validation study in the section on loop substitution in order to show i) how often sampling errors occur in sirtuin side chain optimization, given that side chain optimization is being applied in loop substitution to draw mechanistic conclusions; ii) whether energy errors occur, what kind, and thereafter, consideration of whether such errors may be playing a role in the loop substitution studies.
Later, when we return to part 1 of task list, we will consider similar issues in a very preliminary fashion for loop prediction (where backbone degrees of freedom matter).
RC (4/17): Comments on the latest updates to task list and meeting. Please update task list accordingly.
- The side chain conformation analysis may be presented graphically at first, but later can be quantified using RMSDs.
Note that this analysis will also be relevant to part 3 of the task list where we estimate binding affinities of NAM, since there as well side chains of Phe,Arg may need to be sampled
using rotamer-based optimization. Hence please also comment on the energy difference between the side chain conformations in presence/absence of NAM.
- Note that assessment of accuracy of side chain optimization was part of our plan for assessment of structure prediction accuracy in the context of IFD. We have discussed
the issue of choosing datasets for such validation in the past. Here as well, we should identify some common types of errors and determine whether they are energy or sampling errors (as
discussed during our meeting).
- How long will MM-PBSA single point take to set up? MM-PBSA schedule was not explicitly mentioned in the latest task list
- Ideally MM-PBSA could be set up while completing the next report and could be run immediately after, and included in an updated or separate short report
We can schedule another meeting to discuss that (or review at the very next meeting).
- Ensemble MD calculations on INT complex should be started next and run while moving on to ab initio loop building results analysis.
Note that this should use both MM-GBSA and MM-PBSA.
When reporting results from these calculations, please provide the standard errors as well.
- SIRT3 ab initio loop building results analysis should ideally be done after MM-PBSA, since as noted that problem will not be fully solved in this paper.
Note that as part of this analysis we should include discussion of whether the errors in SIRT3 loop building are sampling or energy errors. Please
provide a high level overview of this issue, indicating in each case whether the native scored better or worse than the refined loop.
Here, the sampling is much more extensive than in the loop substitution tasks, so sampling errors may play an important role. Energy errors
may play a role in both studies. Explanation of causes of energy errors based on inspection of structures generated by ab initio loop prediction is hence also relevant to analysis of scoring
of substituted loops.
Regarding analysis of energy errors, you may look esp at the shorter loop subsections that you have been building, since sampling errors will be less likely here.
- Another meeting can be scheduled to review short report on MD and SIRT3 ab initio loop building analysis
**Please provide some clarity on when these meetings will occur. I have some international travel coming up.
PL(04/03/2015): Updated results for tasks 2.2-2.4 attached to include the side chain optimization results for structures starting from 4BVG. tasks_2.2-2.4_report_updated.docx
RC: Please comment on whether the side chain optimization is leading to energy errors (not just sampling errors). Upon a quick glance it appeared this may be possible as the energies of the side chain
optimized structures after minimization are sometimes found to be higher than those of the structures subjected to only minimization.
PL(04/07/2015): The potential energy surface of protein is too complicated and any restrained optimization prior to the full minimization will probably lead to different minimums. There is no way to determine which protocol will lead to a better minimum. For SIRT3/Int/NAM complexes constructed from 4FVT, it appears that side chain optimized structures do not change the overall energies significantly, probably because the contributing interactions, e.g. h-bonds, hydrophobic interactions didn't change by much when the back bone conformation remain similar.
RC: Regarding the issue of sampling errors, please confirm that the score for the structure prior to side chain optimization and minimization is less favorable than the score for the structure after side chain optimization but before minimization.
Regarding the structural differences, part of the previously assigned tasks was to report the relevant side chain RMSDs. I don't think these have all been reported yet. Please see the task list/wiki and indicate when you plan to report these.
This should include providing the RMSDs before and after minimizations above.
Results for tasks 2.5 (loop refinement for Sir2TM ternary complex) is also attached. tasks_2.5_report.docx
For the task: using loop substituion for the intermediate structure of Sir2TM, which loop should we use? The one obtained using loop refinement for ternary structure (2H4F) or the available loop structure from H116A mutant ternary complex (2H59)? I would suggest we try the loop refinement protocol (with or without distance constraints) on the intermediate complex (3D81) as well for the missing residues (34-44), so we can compare the energies obtained from various methods.
RC: We can use the loop refinement from the ternary complex first and then the available loop structure from the mutant ternary complex.
Yes, we are planning to apply loop refinement to the missing residues in the intermediate complex as part of the Sir2TM tasks. Please indicate here how long that would take and then we will decide whether to proceed with it now.
PL(04/07/2015): It appears that the refined loop from ternary complex will work better because the the loops line up better between ternary complex (2H4F, orange) and Sir2TM/Int (3D81, green), while there is significant deviation for residues 43-47 in mutant ternary complex (2H59, cyan). If we have to replace the loops with the one from 2H59, we will either have to replace a whole loop (residues 34-47), or fix the gap that will occur near residue 43.
For loop refinement that span 11 residues (34-44), it may take one full day to complete the modified ultra extended sampling protocol.
RC(04/08): Ok, we should use the refined structure to start in that case.
I believe you have already started the 11 residue loop building calculation as of today.
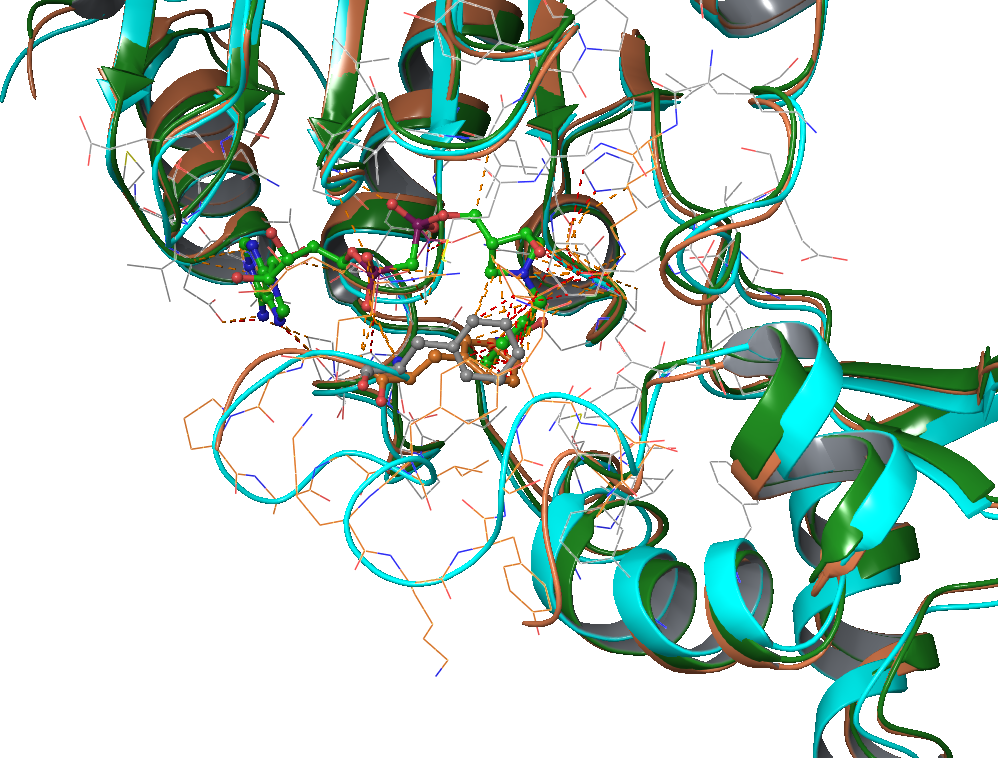
PL(4/2/2015): Update tasks in the schedule file in Dropbox.
RC: The schedule file indicates that the task on MM-GB/PBSA calculations for SIRT3 loops is done. I believe this is done so far for GB only and that PB will soon be underway.
It also indicates that the loop building for Sir2TM is done and I assume we will be moving on to scoring of the substituted loop in Sir2TM. Please provide the results from
loop building and indicate which of the resulting loops will be used going forward.
PL(3/27/2015) Schedule updated in Dropbox.
Report on recent modeling: tasks_2.2-2.4_report.docx. Also available in dropbox.
RC (3/30):
a) Upon a quick review, the following points stand out:
- the h-minimization approach leads to higher energy for the intermediate loop structure starting from 4FVT
- starting from 4BVG we find that the intermediate loop structure has higher energy
As I recall the opposite was found with the previous preparation method. Please comment.
PL: We can't really compare directly the energies for structures prepared starting from 4FVT and those from 4BVG. The later doesn't have ASP395 in SIRT3 and the peptide substrate also differs.
RC: I mean to compare the energy of intermediate loop structure to the ternary loop structure starting from 4FVT.
Similarly, to also compare the energy of intermediate loop structure to the ternary loop structure starting from 4BVG.
PL: For SIRT3/INT/NAM complex structures starting from 4FVT, the energy is lower (more stable, more negative) after loop substitution and Prime full minimization. However, extra step of side chain optimization right after loop substitution can actually disrupt the structure and lead to less stable structures.
For SIRT3/INT/NAM complex structures starting from 4BVG and NAM placement, the native structure has the lowest energy after Prime full minimization, However, early adjustment of heavy atoms may disrupt the structures and lead to less stable structures.
RC: Are you referring to the side chain optimization with the default method?
It appears that for the calculations starting from 4BVG, side chain optimization was not done, but it was done for calculations starting from 4FVT?
Please comment further on the effects of H vs all atom minimization for the calculations with NAM, starting from both 4FVT and 4BVG.(Are the results starting from the two different structures consistent?)
PL: I will run side chain optimization for complex starting from 4BVG as well. As to H vs all atom minimization, it is hard to judge which one is better. Both minimization were done using OPLS_2005 force field only (without solvation model) and restraints were imposed on heavy atoms, therefore can lead to two intermediate stops and different paths in full minimization in the complicated mulch-dimensional potential energy surface. We can choose to use the lowest energies or same protocol for comparison, knowing that there is uncertainty in the final number.
For SIRT3/INT complex, the native structure also has lowest energy.
Could this be due to NAM being improperly positioned? In this regard:
"3. Repeat the above calculations using method 2) proposed below (remove NAM). When substituting the 4FVT loop in 4BVG, this should require no changes to the ligands (intermediate)."
This needs to report full minimization of the native structure (no loop substitution) as well for comparison. It appears only the structure with loop substitution was scored so far.
Also, please repeat this calculation using 4FVT as the starting structure (part of same task).
PL: Full minimization of 4BVG is also included in the following table.
| Struct 13: 4BVG prepared using Protein Preparation Wizard (restrained minimzation on hydrogen only) |
-9620.0 |
| Prime full minimization of Struct 13 |
-12500.3 |
| Struct 14: Replace residue 155-178 of Struct 13 using those of 4FVT (restrained minimzation on hydrogen only) |
-4577.2 |
| Struct 15: Replace residue 155-178 of Struct 13 using those of 4FVT (restrained minimzation on all atoms) |
-11668.8 |
| Prime full minimization of Struct 14 |
-12430.6 |
| Prime full minimization of Struct 15 |
-12496.4 |
RC: Yes
b)
side chain opt: was it applied to only the residues specified on wiki?
side chain opt: please report the side chain RMSDs indicated on wiki, esp for Phe and Arg near NAM, and comment on comparison to minimization only
c)
single-point MM-PBSA: as part of the previous task, do report for both intermediate and ternary loop conformations
ensemble MM-PBSA: as planned for the upcoming task, do for intermediate loop prepared according to the various methods above.
RC (3/27): The task list schedule has not yet been updated per the comments below.
In addition, a progress report indicating what was done this week should also be submitted today.
RC (3/26): The new tasks added to the task list in dropbox do not provide estimated times required.
Please add these today.
Please also indicate which task is currently underway. Are we currently on completion of residue substitution approach using
the 4FVT structure, including MM-PBSA, or have we moved on to residue substitution using the 4BVG structure?
PL(03/16/2015):
| SIRT3/Intermeidate/NAM complex |
Prime Energy (kcal/mol) |
| Convert from 4FVT (ternary complex) |
-12448.3 |
| Convert from 4FVT (ternary complex) followed by loop substitution (res 155-179) |
-12496.3 |
0) Do structure alignment of 4FVT and 4BVG and prepare 4BVG with Protein Preparation Wizard.
1) Start with ternary complex structure 4FVT (SIRT3/carba-NAD+/ac-ACS2): changed carba-NAD+ into NAD+, prepared with Protein Preparation Wizard.
2) Manually create the C-O bond between ac-LYS and ribose, and cut C-N bond between ribose and NAM, use "minimize selected atom" from Tools menu to prepare the SIRT3/intermediate/NAM.
3) Save structures in PDB format and used text editor to copy and replace coordinates of res 155-179.
[The structure obtained above was used in preparation for MD minimization/simulation.]
RC: Which MD simulation?
PL: In part2_of_report.docx, I mentioned some recent MD simulations constructed from parts from 4FVT and 4BVG. The second one mentioned "
SIRT3/Intermediate/NAM complex: constructed from 4FVT after converting carba-NAM/ac-ACS2 to Intermediate and NAM" is what is referred here.
4) Further editing of PDB file to create a separated residue name for NAM, modify the CONECT section. Reload the complex structure w/ and w/o loop replacement.
5) Run Protein Preparation Wizard again, specified the corresponding formal charges on atoms and bonding.
6) Run full minimization using Prime to obtain the final Prime Energies.
RC: Does the above protocol (1-6) correspond to what you did previously? If not, are steps 4-6) and 2) "minimize selected atom" new?
PL: 1-3) described what was done before in preparation for MD simulation. 4-6) was done recently in order to provide an answer to your request. In 2) The atoms selected in "minimized selected atom" include only NAD+ and ac-LYS in order to produce an intermediate under of the conformation of ternary complex.
RC: Ok, then the minimization in 2) corresponds to that proposed in approach 3) below?
PL: Since the ternary structure are not well positioned for the reaction to proceed (unlike Sir2TM), forcing the creation of C-O bond and cutting C-N bond created some awkward bonds that have to optimized. This step is necessary no matter what kind of complexes we want to study.
Does 2) "minimize selected atom" correspond to 3) below (minimize intermediate and NAM coordinates)? In your previous calculations did you also minimize intermediate and NAM coordinates?
There are also several additional questions that were posted earlier below.
PL: In the structure prepared for MD simulation, the minimization for intermediate and NAM was done in NAMD. No further minimization was done in Schrodinger.
I assume 155 and 179 were chosen due to suitably low RMSD, Please provide RMSDs and backbone dihedrals for important residues (including those at loop termini) before and after minimization, and compare dihedrals to those from 4BVG. Previously,
PL: Because I am replacing the coordinates directly using cut-and-paste, the choice of using residues 155-179 ensure no significantly lengthened bonds from backbone atoms (see by-residue CA RMSD in task002.docx)
RC: Ok good, please confirm then that the backbone dihedral angle changes across the loop are negligible upon minimization, and that the only significant changes upon minimization are in side chains (such as Phe157, Arg158)?
PL: The backbone dihedral changes is relatively small upon minimization. ( See attached dihedral calculations for 4FVT, 4BVG, SIRT3/INT/NAM from 4FVT, SIRT3/INT/NAM from 4FVT with loop substitution). The RMSD of the newly constructed SIRT3/INT/NAM complex is 0.593 Angstrom, down from 0.946 for 4FVT. The minimization doesn't cause significant change on the Phe157 and Arg158. A small shift of Arg158 side chain and NAM is found to be sufficient to relieve the clashes.
SIRT3_INT_NAM_complex_loop_reconstructed.xlsx
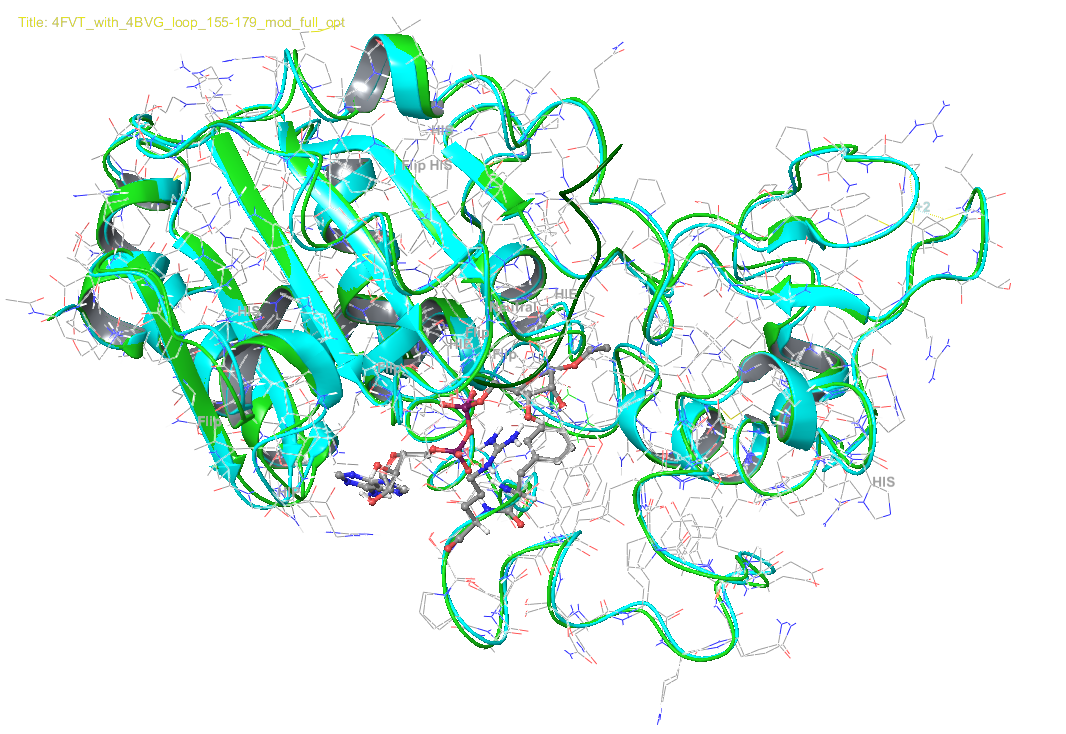
RC: I believe it was stated that there was some issue with structure preparation by loop substitution.
PL: Yes. loop substitution using Schrodinger is creating various issues. That is why I chose to manually edit the PDB files.
Regarding side chains, report 002 indicated that a Phe157 extends into the C pocket while Arg158 remains above C pocket in 4BVG. Does Phe157 clash with NAM in the prepared structure? Was this relieved by minimization?
PL: Yes. Both Phe157 and Arg158 are in severe clashes with NAM and ribose, and can only be relieved after full minimization.
RC (3-18): After replying to the questions posted,
-As noted in the 2nd task in the intermediate loop building section of the task list, please do MM-PBSA scoring of the complexes as well.
PL: I am not sure if this is what you expected. Here is what I got in calculating the Prime energy (obtained with VSGB solvation model): All structures starts from Step 5 mentioned above after another round of protein preparation wizard. The first two keep the position of all heavy atoms (therefore heavy collision after loop substitution.) and minimized only the hydrogens using OPLS_2015 force field. The next two were optimized within Protein preparation wizard using restraints and OPLS_2015 force field. (without solvation). The last two were optimized using Prime, which does use VSGB solvation model.
| SIRT3/Intermeidate/NAM complex |
Prime Energy (kcal/mol) |
| Convert from 4FVT with optmization on proton only (OPLS) |
-10186.1 |
| Convert from 4FVT followed by loop substitution (res 155-179) with optmization on proton only (OPLS) |
111172617.0 |
| Convert from 4FVT with renstrained optmization (OPLS) |
-11914.0 |
| Convert from 4FVT followed by loop substitution (res 155-179) with restrained optmization (OPLS) |
-11925.7 |
| Convert from 4FVT (ternary complex) with full Prime minimization |
-12448.3 |
| Convert from 4FVT followed by loop substitution (res 155-179) with full Prime minimization |
-12496.3 |
PL:(03/20/2015): No, there is no option in Schrodinger to use PB other than the VSGB method.
RC: Ok, we can then do MM-PBSA calculations in Amber.
-As part of the 2nd task, please run side chain optimization for residues on the loop and within 7.5A of the loop for both structures prior to the full minimization step and report the energies after side chain optimization and minimization. Also report the RMSDs of the side chains in the structure (on the loop and its environment) with loop substitution to those in 4BVG. Here I am assuming the backbone RMSDs are negligible (see my question above).
PL: There is "Predict Side Chains" module available in Prime. Which structures do you want to apply the method to, considering that we have different complex structures optimized at different levels? Do you want to cover all the residues 155-179 plus those within 7.5A? One issue with the side chains optimization is that the coordinates of NAM and intermediate may have a significant impact and they are not optimized in some of the structures above.
RC(3/20): We should apply it to the "Convert from 4FVT" and "Convert from 4FVT with loop substitution)" complexes, after the initial minimizations for ligand preparation but before the full minimizations. The full minimizations should be carried out after side chain optimization. We should apply it to the residues you mentioned above (later, we can consider doing another side chain optimization on only the loop residues). It is important to do side chain optimization because, e.g., Phe and Arg clash with NAM, and other side chains in the loop environment may be different for the 4FVT vs 4BVG (intermediate) structures.
PL:(03/20/2015): The order of optimizations has significant impact on the final result in the complex built with loop substitution. The original construction of intermediate and NAM from 4FVT was not very robust (didn't assign new resname for NAM may cause some issue with the optimization, and not re-assigning force field parameters may raise question about the energy associated. A save-edit-reload-reassign-re-optimization procedure is probably needed to ensure the quality of the result.). I will revisit the procedure and make sure all details were taken care of.
RC: Ok that is fine; please first update the schedule as noted according to the 3/20 comments and then proceed with this.
Ab initio loop predictions use the above protocol for side chain optimization, and hence we use the same method here (the loop backbone is already optimized).
-Also, as an alternative method of preparing the complex, substitute the 4FVT loop in the 4BVG structure, preparing NAM by alignment, and compare the energy difference.
The part of the task pertaining to low energy loop conformations from prime can be omitted for now.
RC (3/20): A new task should be added below "Completion of residue substitution approach to intermediate loop building"
in the task list: repeat the above calculations using method 2) proposed below (remove NAM).
When substituting the 4FVT loop in 4BVG, this should require no changes to the ligands (intermediate).
In pt 1 of the task list, there was a postponed task related to filling in "missing loop residues of Sir2 once we are comfortable with refinements". Next, the part of this task relevant to our current work can be revisited: applying ultra extended sampling to build missing residues in the Sir2Tm ternary complex only (fewer missing residues). The rest of the task can be postponed. We may return to the intermediate loop conformation (11 missing residues) after further work on accurate loop prediction validation in 2H59.
Then, another new task pertaining to Sir2Tm should be added below "Completion of residue substitution approach to intermediate loop building": score the ternary loop conformation (MM-GBSA and PBSA) in the ternary and intermediate xtal structures for Sir2Tm in a manner analogous to that for SIRT3 (using loop substitution for the intermediate structure), but using ultra extended sampling to build the missing residues in the ternary loop.
Include side chain optimization as above.
Please add these tasks to the task list document and provide estimated times for each, as well as updated time estimates for the earlier SIRT3 tasks underway (this is no longer a half day task).
PL (03/13/2015):
I start to work on the SIRT3/Intermediate/NAM using 4FVT with loop (residue 155-179) from 4BVG. In this setting, the original intermediate and NAM coordinates are not optimized. Therefore, we have to a few options to investigate the overall energies w/ and w/o the loop substitution: 1) remove intermediate and NAM and minimize the structures for comparison; 2) remove NAM and minimized intermediate first before minimize overall structures; 3) minimize intermediate and NAM coordinates first, followed by minimizing overall structures. Please advice what is the most appropriate option for your needs.
RC: Previously, you indicated that an attempt was made to prepare this structure with loop from 4BVG. The first task in section
"Building of intermediate loops starting from ternary loop conformation" requested information on the previous attempts and the results obtained, including
any issues observed with matching of loop termini.
PL: Yes. I have the structure for the complex prepared before and used in MD simulation to calculate binding affinity.
RC: Please provide the details of how the earlier complex was prepared starting from 4FVT with loop from 4BVG (which ligands were present, how they were prepared, etc), the structure prepared, and how you assessed the quality of the results of the structure preparation (e.g,, any issues observed with matching of loop termini). There was also a question regarding the loop dihedral changes; if these did not change at all, it appears that no minimization was carried out prior to MD?
I think you want to use Prime to calculate the overall energies before and after the loop substitution and compare the energies. If that is true, what I mentioned above is relevant because the intermediate/NAM coordinate is not optimized in the prepared structure, and we need to choose how to deal with the intermediate/NAM structure before calculating the overall energies.
RC: Yes, that is one of the goals. We should include both NAM and intermediate in our first attempts to build a model. Hence we should ideally start with approach 3) after providing the info requested above on the previous results.
As I interpret options 1 and 2, they appear to remove both intermediate and NAM, or only NAM, respectively (2 should read "remove NAM and minimize intermediate first"?).
In task 2 report, it says that the flexible loop region can be identified as between residue 155 and 174. Why 179?
Given the potential issues with structure preparation of the two ligands, before proceeding with this 2nd task in intermediate loop building section, we will enumerate in detail the planned steps for preparation of the SIRT3/Intermediate/NAM, including how the starting structures of intermediate, NAM are being prepared. I believe 4FVT contains carba-NAD, whereas 4BVG contains intermediate. I will comment further on this after receiving the results from the first task.
PL(3/12/2015): Prime energies for various structures of 2H59 in Loop refinement.docx
The 2H59 loop refinement validation test seem to suggest that the prime loop refinement using modified ultra extended sampling protocol works well for less than 10 residues because no near native structures can be found for residues 36-45 or 35-46 (by visualization, further analysis will be provided.)
The Prime loop refinement jobs are currently set to run on Windows Desktop for consistency because the Linux version produced different results for the same input file.
RC: ideally.we can run jobs on Linux going forward. How are you submitting jobs in queue?
Although multiple jobs can be submitted at the same time, which I often did, the result can take longer to obtain. A 12-residue loop refinement with modified ultra extended sampling takes about 2 days to complete, and it may request 18 tokens at the same time as the number of subjobs launched (the specified maximum subjobs in the log file for such loop refinement job is 289).
Last Friday, I submitted three loop refinement jobs for SIRT3 systems. Two of them did finished, and one failed, citing failed to check out license in one of its subjobs.
The two finished jobs for SIRT3 systems are for 14-residue loop refinement (156-169) and no near native structures can be found by visualization.
RC: Given the time required for jobs to run, it is stated below (and in several previous emails) what work is to be carried out while the jobs run - after jobs are submitted in queue, not after all the results are obtained. This is an issue of time and resource management given the priorities and deadlines of the company (in this case, progress on an important calculation). We should be on the same page about such time management in advance. In the future as well, if jobs are to take several days to run, please let me know so we can schedule other work in the interim. Just as with lab work, computational work should be done in parallel to the greatest extent possible, with constant attention to maximization of productivity.
RC (3/6): As specified in my emails starting Tues, it is important that progress is made on residue substitution tasks in Section II of the tasks list
by next week. In order to achieve this, it was indicated that loop prediction jobs should be prepared and submitted (in batch if required), and while
jobs run, to make progress on the first residue substitution task. In order to ensure that the plan of work can achieve our goal for next week,
I asked on Tues for a schedule for this week and next week. The schedule proposed does not appear consistent with this goal:
-The schedule posted today indicates that 2H59 jobs will take 4 days. From which date?
It is stated that complete analysis is not yet ready. I would like to see a brief summary of the results today.
-The schedule indicates that setting up and running SIRT3 will take 6 days (this does not include analysis of results). The time
allotted should specify the number of days required to set up and submit the jobs, not the time required for the jobs to complete.
I assume 6 days includes the time it will take for the jobs to complete. When will they will be submitted? Please clarify.
-As noted, I would like to know what will be done each day next week so I can verify that we will have made sufficient progress on the residue substitution
tasks by next week.
PL(03/04/2015): loop backbone RMSDs for the residues predicted.
summ_full_RMSD_sets12.xlsx (see the additional lines marked as RMSD(loop))
PL(02/26/2015): Comparison of Prime Energies of selected 2H59 structures. Comparison of Prime Energies of various 2H59 structures.docx
A compilation of Structures, Full RMSD, Backbone RMSD, RMSD for residues 33-47, Prime Energies, by residue RMSD (full residue or backbone only).
summ_full_RMSD_sets12.xlsx
summ_bb_RMSD_sets12.xlsx
RC: Please provide commentary accompanying the data provided and also present answers to the latest questions posed on the wiki below regarding consistent energetic comparison, explaining how this data relates to each subtask for the current task in tasks doc so we can assess how much of this task is complete. Please also describe the resolution of the issues you mentioned below. Please define all the terms - including "matching side chain after step 1".
PL(02/27/2015): The " + 7.5 A matching side chain " described in the minimizing run means that the side chains included in the loop refinement are hand-picked so that the overall movable atoms are consistent between the minimizing run and loop refinement run, which presumably make the energetic comparison meaningful.
As to the results, for loop 37-44, the predicted loop structure is no better. Remember that the reported backbone RMSD for residue 33-47 is ~ 2 A, which was significantly lowered by not having contribution from original residues at 33-36, 45-47.
RC: Please report the loop backbone RMSDs for the residues predicted (e.g. 37-44) for each length loop.
Also, there appears to be some overlap between the data in the two xls above. Please clarify which data are different.
PL(03/04/2015): The top few lines are the same between the two files. The difference is on RMSDs for each residue, one uses all atoms (full), the other uses backbone atoms only (bb).
As you have suggested, we might need to explore the ultra-extended sampling for 35-46, or even 37-44 to help draw conclusion of the sampling or potential that need to be improved. I will need to look at their original paper to comment more on the results.
In 2007, there is a evaluation and comparison done by researchers from BMS, which shows that although Prime performs best compared to other tools, it can fail for medium length (8-10) loop prediction.
loop_refinement_1999.pdf
RC: I previously reviewed the latest long loop prediction papers upon which the ultra extended sampling methods are based and they have improved accuracy beyond that reported in the review above. In particular, accuracy is reasonable for loops above 12 residues using these methods. There have been improvements in both the energy function (VSGB2) and sampling methods that have helped achieve this. I will comment on this further later including next steps in this regard for our longer loops after getting some additional results below.
In the meantime, the following should be done next:
-specify how the energy of native changes when the same side chains in the loop environment that were optimized in the loop prediction are optimized in the native as well.
-set up/run (batch) jobs for 2H59 loops lengths you mentioned above (37-44,35-46) and also for SIRT3 ternary and intermediate complexes with ultra-extended sampling and more fixed stages (multiple lengths), for purpose of further loop prediction validation.
-move on to residue substitution task 1 - i.e., the one that ask for explanation of what was done previously on residue substitution after alignment
Please update the task list accordingly including estimated time needed for setting up these jobs (not completing or analyzing the results) and the preliminary residue substitution task. Please specify in the task list what loop lengths you are looking at.
It is important to move on to the third task above shortly, so I would like to have an idea of the time required.
Upon a brief review, the following observations are made (referring to backbone RMSD for the loop residues in the xls):
- for 37-44, we appear to have acceptable predictive accuracy compared to native with RMSD within reasonable limits (although perhaps not as good as other representative results reported in the literature for this length). sampling does not appear to be a major issue, and the native scores better than the model as expected - suggesting some sampling error is contributing.
- for 35-46, which is a (super) long loop, accuracy is compromised, but the sampling is only at the extended level, and the energies of the models are higher than that of native with matching side chains, so this may be a sampling error. This could potentially be validated in the next step with ultra extended sampling with fixed stages.
- for 33-47, we see significantly lower accuracy, and if the method of energetic comparison is acceptable, we find that the ultra extended sampling with 10 fixed stages identifies a model with lower energy than native according to the energy function despite significant structural differences. This becomes apparent only at the level of 10 fixed stages, not 5 fixed stages. This may be indicative of an energy error for the super long loop. Again these conclusions would depend on assessment of the consistency of the energy calculation methods.
These comments are based on a very brief review of the dataset. Please expand upon these observations with references to more of the information in the dataset (including other length loop predictions).
Please also indicate which of the loop lengths are relevant to the problem of filling in the missing loop residues in Sir2Tm (from reports below).
Please provide the above replies today to enable planning of immediate next steps in the tasks doc.
PL(02/24/2015): Updated tasks schedule is available on Dropbox/PMC-AT PLIN/Computational_tasks_schedule_phase_I_2-10.docx.
PL(02/18/2015): partial results of 2H59 loop refinement test.
CA_RMSD_2H59_results.xlsx
Notation used in the Excel file:
2H59_ref: 2H59 prepared using protein preparation wizard and minimized using OPLS_2015 with constraints on heavy atoms (RMSD < 0.3).
full_min: minimization using Prime.
min_37-42/min_37-44/min_35-46/min_33-47: minimization of residues in the range plus residues within 7.5 A.
lr_37-42_1,..... : lr: loop refinement using extended sampling; 37-42: for residue in between 37 and 42, with side chain within 7.5 A optimized. _1: ranking out of the 10 outputs.
lr_33-47_ue_1, ......: loop refinement using ultra extended sampling.
lr_33-47_mue_1, ......: loop refinement using ultra extended sampling plus 5 initial stages
lr_33-47_mue2_1, ......: loop refinement using ultra extended sampling with 5 initial stages and 10 fixed stages.
The CA_RMSD is actually the CA distance between two models.
Some clarification:
1) The energy -11137.7 and -11024.5 is not comparable, because 1) the loop refinement covers shorter range of residues in definition (34-46) vs (33-47) in the minimization; 2) only side chains of the residues within 7.5 A are optimized in the loop refinement. I am working on some new calculation that will be more comparable.
2) Although 10 structures were generated from loop refinement, many of them have little differences.
3) No further alignment is done in the calculation of the CA_RMSD.
PL(2/25/2015): The residue defined in loop refinement is actually what is defined, not shorter range: i.e. res 33-47 do mean 33-47, not 34-46. However, the backbone change is usually very small (< 0.08 Angstrom), which causes me to mistaken it as not changing.
The residue side chain selection within 7.5 Angstrom differ between loop refinement module and prime minimization selection, with later being larger. Probably the side chain has to be all within the distance range to be selected for loop refinement, while the prime minimization selected all side chain with any atom in the range.
I will run some minimization using the same residues to make them comparable. Results will be posted accordingly.
Here are some more results:
| Sir2TM H116A mutant ternary complex loop refinement |
MM-GBSA |
RMSD (heavy atoms) |
RMSD (res. 33-47) |
| step 1: protein preparation wizard |
-10964.2 |
||
| minimizing res 37-42 + 7.5 Å after step 1 |
-11041.8 |
0.3174 |
1.2689 |
| minimizing res 37-44 + 7.5 Å after step 1 |
-11053.9 |
0.3496 |
1.3566 |
| minimizing res 35-46 + 7.5 Å after step 1 |
-11087.4 |
0.3797 |
1.1943 |
| minimizing res 33-47 + 7.5 Å after step 1 |
-11137.7 |
0.4351 |
1.3338 |
| full minimization |
-11634.6 |
1.3626 |
1.6938 |
| loop refinement on res 37-42 after step 1; extended sampling |
-10999.6 |
0.3231 |
0.6845 |
| loop refinement on res 37-44 after step 1; extended sampling |
-10989.7 |
1.1314 |
5.0057 |
| loop refinement on res 35-46 after step 1; extended sampling |
-10981.3 |
1.379 |
6.2297 |
| loop refinement on res 33-47 after step 1; extended sampling |
-10982.2 |
1.8951 |
7.8548 |
| loop refinement on res 33-47 after step 1; ultra extended sampling |
-10998.6 |
2.9828 |
12.5306 |
| loop refinement on res 33-47 after step 1; modified ultra extended sampling |
-11005.1 |
1.4036 |
6.2462 |
| loop refinement on res 33-47 after step 1; modified ultra extended sampling, 10 fixed stages |
-11024.5 |
2.1737 |
8.6814 |
RC (02/19/2015): To communicate and plan effectively on the remaining items for this task (code for processing and backbone RMSD calculation of output structures, proper comparison of energies as noted above). please separate them from the items completed so far on this task in the task list and provide estimated time required; RC (02/23): This (along with answers to the specific questions below) should be posted asap.
PL (02/24/2015):
-can we choose more than 10 models to report?
PL: Yes.
-in the xls, it appears that none of the models were subjected to full minimization. please confirm
PL: On the table, there is only one from the original structure after protein preparation wizard got fully minimized. There is another model from loop refinement got fully minimized by not included in the report.(It has a total Prime energy of -11611.9 kcal/mol, higher than the -11634.6 of the minimized original structure.)
-"loop refinement covers shorter range of residues in definition (34-46) vs (33-47) in the minimization": this loop prediction is described as res 33-47 above; I assume (34-46) refers to the minimization of loop residues after the loop prediction but not the residues 33-47 sampled in the loop prediction. It also appears from comments above that you are doing side chain optimization but not minimization on residues within 7.5A. Please confirm or clarify.
PL: No, in reverse. The loop refinement, a selection of res 33-47 lead to changes of backbone atoms on residues of 34-46. However, the residues selected for minimization will include completed res 33-47.
-Regarding new comparable calculations, please refer to the notes on this provided in the task list and indicate which of these are being done / how the new comparable calculations relate to these
PL: The compatibility between minimization and loop refinement is at the issue with the definition of selection, definition of range for within 7.5 A, and the optimization method for side chain. I am able to make selection in the minimization to include only sidechains within 7.5 A, and it appears there may still exist some discrepancy between the selection. I am still testing and make comparison about the difference, and will update accordingly.
RC: Is side chain optimization for loop prediction carried out for sidechains within 7.5A of the model loop? By making selection in the minimization to include only sidechains within 7.5A, do you mean within 7.5A of the native loop conformation? Please note that since the loop is being built in non-native conformations, the residues being optimized and minimized under the energy function may differ between native and model loops. (This could be assessed for the models reported in the xls.) Regarding optimization, the native structure results above were only minimized not optimized. See comments/approaches related to dealing with this in task list. Please comment on other issues related to the discrepancy that are being tested.
-will wait for the backbone rmsd and proper energy comparison before providing further comments, but prediction accuracy for even 37-44 (8 residues) may be low given the data above. Sampling should be better at this length; hence this could be an energy error rather than a sampling error - this should be validated upon proper energetic comparison.
PL: Coding for calculating RMSD using either backbone atoms, by residue, selected range of residues, overall rmsd, etc. is mostly completed and working fine.
As to the error for shorter loop (37-44, actually it is sampling backbone of 6 residues from 38-43), I am not sure it is the energy error. It is more likely that the initial prediction fail to produce near-native conformations.
RC: Ok. We should be able to validate this when our compatible energy calculations are ready. We can use a higher level of sampling if needed to check if we can solve the problem for shorter loops.
PL (02/16/2015): A quick update on loop refinement results:
| Sir2TM H116A mutant ternary complex loop refinement |
MM-GBSA |
RMSD (heavy atoms) |
RMSD (res. 33-47) |
| step 1: protein preparation wizard |
-10964.2 |
||
| minimizing res 37-42 + 7.5 Å after step 1 |
-11041.8 |
0.3174 |
1.2689 |
| minimizing res 33-47 + 7.5 Å after step 1 |
-11137.7 |
0.4351 |
1.3338 |
| full minimization |
-11634.6 |
1.3626 |
1.6938 |
| loop refinement on res 37-42 after step 1; extended sampling |
-10999.6 |
0.3231 |
0.6845 |
| loop refinement on res 33-47 after step 1; extended sampling |
-10982.2 |
1.8951 |
7.8548 |
| loop refinement on res 33-47 after step 1; modified ultra extended sampling |
-11005.1 |
1.4036 |
6.2462 |
| loop refinement on res 33-47 after step 1; modified ultra extended sampling, 10 fixed stages |
-11024.5 |
2.1737 |
8.6814 |
- The protein preparation wizard involves the following steps: 1) pre-processing: Assign bond orders, add hydrogen, fill in missing side chains, cap termini, delete waters beyond 5 Angstrom of het groups. 2) Run Epik for het groups. 3) Run interactive optimization of H-bond network and protonation states. 4) Remove waters with less than 3 H-bonds and run restrained minimization (in this case, with constraints on heavy atom to RMSD < 0.30 Angstrom. sometime, minimization carried out for hydrogen only.)
- modified ultra extended sampling run 5 initial stages with the specified minimum overlap factors instead of 3 initial stages, which allow more initial structures to be generated.
- modified ultra extended sampling, 10 fixed stages also increase the number of fixed stages to 10.
RC: See comments on task list regarding proper comparison of energies of predicted and native loop conformations. Are the structures with energy -11137.7 and -11024.5 comparable in this regard, with respect to side chains optimized and residues minimized?
See also comments about reporting of data on multiple predicted low energy loop conformations. (Also indicate how many loop conformations are provided in the prime output so I can assess how many of the prime-generated loops you examined when choosing those with low RMSD to native.)
Note the type of RMSD data requested in the task list.
Please provide the report for the parts of this task completed so far with these points included. As noted, next steps may be affected by the results and some lead time will be needed for review, so please provide the above info as it becomes available.
RC (02/13/2015): Please provide an update on 2H59 loop modeling task today with results available to date, to facilitate planning of next week's tasks. Thanks.
PL (02/11/2015):
Residues in contact with NAM.docx
PL (02/10/15): modeling of 2H59 loop structure is currently underway. tasks schedule file is updated and available under /Dropbox/PMC-AT PLIN/.
Can you clarify what you have in mind on comparing the contacts with NAM for ternary and intermediate complex? The xtal structures of intermediate complex do not have NAM in the pocket. If preparing intermediate complex with NAM from ternary structure, there is usually little change in contacts with NAM.
RC: For example, Phe33 (Sir2Tm) and perhaps Phe157 (SIRT3) blocks the C pocket after intermediate formation (and NAM dissociation).
Perhaps there is no relevant contact to NAM prior to dissociation.
In that case, are there any other conformational changes upon intermediate formation that involve changes in contacts to NAM?
Wolberger mentioned that there may be an NAM-flipping event that presumably would lead to changes in contacts. However, this may have been speculation; I believe we considered it earlier.
Related, you noted in task002 that Arg158 (SIRT3) sits above the C pocket. Does this occur only after loop conformational change, and does it involve any new contacts to intermediate?
The steps involved in protein preparation wizard has been described by Eric before in Protein.Preparation.Protocol.README.doc (under Eric's maestro.projects.and.simulations directory). My protocol is similar to his and will provide a detailed description later.
RC: The summary of NAM interactions indicated that there are important differences in Phe157 conformation in ternary and intermediate structures. "The residues are consistent in ternary complex and intermediate complex except for PHE157 in 4FVT, which is not in contact with NAM and highlighted with yellow in the table above." Does this mean Phe157 is not in contact with the NAM moiety of NAD+ in the ternary complex, but that it occupies the C pocket in the intermediate (BVG) complex? Is the situation similar for Sir2Tm (the descriptions and annotations of structures were more detailed for SIRT3)?
PL(02/10/15): Task list file is now placed under /Dropbox/PMC-AT PLIN/, which I will update frequently.
RC (02/09/15): Updated task list.
Computational_tasks_schedule_phase_I_2-10.docx
RC (02/10/15): In order to access the latest sampling algorithms (dipeptide sampling with fixed stages) for long loop prediction, which can improve accuracy for 14-20 residue loops, it may be necessary to run prime loop prediction from
the command line with manual preparation of the input file. They may not be available from the gui. Please for see http://www.schrodinger.com/kb/1475 for needed input file commands. Fixed stages refer to sampling stages where a specified number of residues at the ends of the loop are fixed while the remainder of the loop is sampled (more fixed stages may be required for even longer loops).
The initial stage of the algorithm uses 5 different steric overlap factors (indicated in the input file) to build starting conformations. A refinement stage constrains the Calpha atoms for more focused sampling. Then come fixed stages, followed by another refinement stage.
Tasks schedule above has been updated accordingly. Use of other features may also require running from command line (see updated notes in Task003 doc below).
Please indicate when the 2h59 loop prediction task is underway and let me know if you are able to access this feature. As noted in the task we would like to establish whether there are any energy errors.
The loop sampling may take longer; if so please update the estimated time required for the task.
PL(02/06/2015): follow-up on task 1 & 2.task001_PL_RC_v1.docxtask002_PL_RC_v0.docx
task001_PL_RC_v2.docx
RC (2/04/15): Please reply to follow up questions on tasks 1-3 before proceeding with 4 (some answers may affect subsequent tasks).
For the longer questions (esp under task 3), you can add the task to the task list under task 3 and before task 4.
PL: (02/05/2015): updates to task003 alignment2.pptx
RC: Further comment on task003:
Task003_PL_RC_v3.docx
Alignment with XG's annotations on catalytic residues has been placed in PMC-AT PLIN dropbox.
PL(02/04/2015):Task003_PL_RC.docx
RC: Follow up questions on task 3.
PL(02/04/2015): task002_PL_RC.docx
RC: Follow up questions regarding task 2 above.
PL(02/03/2015):
task001_PL_RC.docx
RC: Some follow up questions regarding missing loop residues added to doc above.
task list and required time.
Computational_tasks_schedule_phase_I_PL.docx
RC: Please see 1) below. Order of Phase I tasks has been provided (start top of pg 1 and proceed sequentially down unless we specify otherwise in advance). Please work in the order listed. Some of the earlier tasks are essential for proper communication and planning of subsequent work: e.g., the first two tasks on reasons for choice of loop residues, and later "Providing more details on results obtained to date with residue substitution approach to intermediate loop building including dihedral comparison after MD".
I.e., listed status of tasks #1, followed by #2, etc on page 1 of doc should be "underway" at this time and status of each task should also be updated when completed (see 4) below)
Please let me know if you have any specific questions on tasks.
RC: Section "Building of intermediate loops starting from ternary loop conformation" refers to building the intermediate loop starting from the the ternary receptor xtal structure (4FVT). I believe the tasks you have reported on last Fri pertained to validation of loop prediction by rebuilding the intermediate and ternary loop conformations within their respective native environments or under a change of carba-NAD to NAD. This can go in section I of phase I next to the other validation task - please add the task (e.g. next to the 2H59 validation task) and indicate it is complete. (I will provide feedback on these results shortly while progress is made in order starting from the first couple of tasks on pg 1.)
RC: You appear to have misunderstood the intended format of the table "Computational_tasks_schedule_phase_I".
"Tasks listed 1/28, esp validation of loop energetic scoring and sampling, Sir2 and SIRT3" (in bold in the table) is a section header like the other
boldfaced text in the table, not a task. The tasks listed 1/28 were reorganized and listed below it in the table (as will be clear from reading those tasks under the header in the table).
Estimated days required for all phase I tasks (that is all tasks currently in the table under the various bold headers) should be provided by EOD Mon. Please provide directly in the table "Computational tasks schedule phase I.docx".
PL(01/30/15): Investigation of constrained loop sampling to build the intermediate loop starting from the ternary conformation, in both Sir2Tm and SIRT3: part 3: SIRT3.
part9_of_report.docx
I just finished summarizing my previous work, and begin looking into the new tasks. I will update the status as soon as the task is underway.
RC: After reviewing the table, please provide estimated number of days required for each task in the appropriate table column as well (see #2 below).
This information should be posted on Mon morning prior to any other work.
Most of the tasks were previously assigned but are now organized in desired order of priority.
RC (1/29): List / schedule of tasks (phase I) previously mentioned is provided below. It will be updated with more details on an ongoing basis by both PL and RC.
1) order of Phase I tasks has been provided
2) we have a sense of an appropriate timeline for Phase I given the publication strategy and time that will be required for Phase II tasks (also required for this paper). This Fri, please propose estimated number of days required for each task and RC will comment/edit as needed. (Number of days refers to the task description as written, not additional follow up tasks that may be needed based on results from those tasks.)
3) when you start work on each task, please update its status to "underway"
4) as soon as you complete a task, update its status as "completed" and provide results/next section of report on wiki before moving to the next, so RC can analyze and provide feedback / additional planning as needed before we move on.
Computational_tasks_schedule_phase_I.docx
PL(01/27/15): Investigation of constrained loop sampling to build the intermediate loop starting from the ternary conformation, in both Sir2Tm and SIRT3: part 1: Sir2TM.
part8_of_report.docx
RC (1/28): 1/28 comments (here and below) focus on further information required regarding loop building tasks that is necessary for advance planning of the upcoming simulations.
- latest plans for loop building in SIRT3 suggest that a subsection of the loop is most flexible and will be built for purposes of upcoming simulations, whereas the early
loop building tests (in the native environment) considered the whole (>20 residue) loop. Validations of ab initio loop prediction should thus be carried out for the flexible
subsection of the loop.
- for example, Sir2Tm 2H59 binding loop should be built with and without analogous constraints to those used in report pt 8, and by-residue RMSDs should be reported. For this, results for several of the highest ranking
loop structures can be provided, since the highest ranking structure may not be the one with lowest RMSD. The energy of the native loop (with/without minimization) should be included for comparison.
- loop refinement for 3D81 (intermediate) showed the constrained sampling can lead to energy improvements and hence that the MM-GBSA scoring may be capable of accurately (more information in this regard will be provided by the previous 2H59 validation above).
- similarly, for SIRT3 4BVG, the distance constraint matrix below should first be applied to rebuilding of loop in the native 4BVG environment and analogous data should be provided.
- please indicate whether the refined loops for 2H4F contained a helical segment (see also below) in the case that the sequence alignment suggests that the helical segment should lie within the range of missing residues (if not could specify helical constraint, if supported, to check the energy difference and validate sampling)
- specify in structure-based sequence alignment (meetings page) where the binding loop is and indicate the residue numbers for the loop residues for SIRT3 and Sir2Tm with
numbering consistent with the reports
- also highlight the helical segment in the ternary structures (e.g., 4FVT) in this alignment
- also indicate the residue in SIRT3 that is homologous to Phe33 in Sir2Tm. Is it Phe157 (noted from pptx below). Please do the same for Tyr40. PL(01/30/15): Yes, PHE157 in SIRT3 corresponds to PHE33 in Sir2TM, TYR165 in SIRT3 corresponds to TYR40 in Sir2TM.
PL(01/22/15): Part 5 and part 6 of the report. part5_of_report.docx part6_of_report.docx
The summary of the binding loop conformation investigation in literature and crystal structures. Sirtuins reaction mechanism & related structural changes.pptx
RC (1/28):
- this report indicates there is a helical segment (162-170) within the binding loop for the ternary complex of SIRT3. It is not clear whether there is such a helix within
the resolved loop residues of Sir2Tm and if so, where; this should be specified PL(01/30/15): there is indeed another Sir2TM complex structure with the loop resolved and there is a
3/10 helix in the corresponding position. I will summarize.
And here are some answers to RC's questions. some_answers_to_RC_questions..docx
PL(01/09/2015): Part 4 of the report part4_of_report.docx.
RC (1/28): By reference to the by-residue RMSD figure, please indicate a) the loop residues; b) choice of residues for intermediate loop building in Sir2Tm.
Also please zoom in on the loop residue numbers in the x-axis of these figures (omit more distant residues).
PL(01/08/2015): Part 3 of the report part3_of_report.docx, and associated data file for CA-distance matrix and by-residue RMSDs CA_distance_matrix_and_by-residue_RMSDs.xlsx.
RC (1/28): By reference to the by-residue RMSD figure, please indicate a) the loop residues; b) the reason for choice of 156-169 for the intermediate loop building starting from the ternary structure with constraints.
Also please zoom in on the loop residue numbers in the x-axis of these figures (omit more distant residues) so the residue numbers are clear.
PL(12/26/14): Part 2 of the report part2_of_report.docx, further updates will follow.
Associated files: MMPBGBSA_data_summary_v2.xlsxresults_12-26-2014.xlsx
PL(12/12/14): Part 1a of the report, part1_of_report.docx.
RC: Based on our discussions (wherein we asked for an status update on all computational tasks currently underway by Fri) it is not clear whether this will be followed by another report (Part 1b? Part 2?) on Mon.
Regarding Pt 1(a):
- are MM-PBSA calculations underway? please provide when ready. you may also comment further on the studies you previously did on the correlations between MM-PBSA, GBSA, glidescore summarizing the observations from that study
- did you carry out MD sampling yet? previously, this did not make much difference, but if the new molecules are larger than those from the NAM analogs, it could. please comment
- please report R^2 from linear regression of each type of computational score against the experimental data using the entire dataset (both the new molecules from the literature as well as the old molecules we previously studied in our lab)
- after receiving all these I will look further into the next steps discussed in our email on this topic (including the range of binding affinities represented vis-a-vis the standard errors)
- The kinetic modeling will be used alongside computational modeling (c pocket binding affinity correlations) to estimate some important parameters including kd,nad and kex. Xgs draft has some of the models. I'll describe more later.
- see below regarding loop MD
Given the recent feedback and questions provided on the wiki:
a) Several tasks identified as being particularly important to the paper were not covered in Part 1a. E.g., building of the loop in the intermediate complex starting from the ternary structure,
and the associated energetic analysis. Please update on these asap.
b) It was indicated below that continuation of MD simulations started previously is not included in the list of priority tasks relevant to the paper writing, since these take little time to set up and especially to monitor (questions were also posed on the cpu capacity usage by MD simulations)
c) Other questions have been posed recently on the wiki pertaining to the paper. Please answer these. A question was also posed to XG in the context of the C pocket binding affinity studies with regard to the standard errors of the experimental measurements
e) Status update should also be provided (with XG) on the focused computational/experimental literature analysis that was outlined for the purposes of the paper.
In general you need to describe in detail how your computational results align with the state of art of literature knowledge regarding the relevant parts of the sirtuin mechanism described in the tasks. As you can tell the emphasis is on the nam cleavage and immediate next step. Energy differences are being studied computationally in the tasks.
f) It is not clear which other tasks are currently underway. Please update the tasks_list.docx below at the start of the week to indicate which tasks (other than continuing MD simulations) are underway and their status.
g) Please provide your estimated schedule for the next week (including next steps for each of the tasks underway).
h) Please ensure that AS provides his schedule for the next week as well. I will reply separately to your email regarding MM-GBSA code development. Since that requires some further planning, his schedule for this week should not depend on it.
PL(11/26): The task list is rearranged by date and status in a separate column. task_lists_updated.docx
Two reports of some recent work: Loop refinement for 2H4F_4FVT.docx Partial summary of MD simulations.docx.
There are also files saved to the Dropbox.
RC: if carba nad causes an issue w loop conformation in sirt3, what about in corresponding sir2 structures used for qmmm simulations? have you compared the structures?
RC:
- i will review the movies
- which BVG-prepared structures show smaller B factors? do you mean using BVG to prepare the intermediate in the 4FVT protein structure?
PL(12/02): What I referred to is the simulations started with SIRT3 structure taken from 4BVG/4VB3/4BVH, which bear similar loop structure, and were found to be stable during the course of the simulation.
RC: This is not surprising given that these are the intermediate complex xtal structures; I believe this was already mentioned earlier.
Please consider the following comments as constructive feedback on the progress/status updates:
- there has not been sufficient progress shown toward milestones that can be used to write the paper
- priorities in this regard: tasks other than completing the md simulations. md simulations are easy to set up and as you know, take a long time to run.
- you need to connect up with xg and make sure the tasks on computational and experimental literature review on sirtuin mechanisms are completed soon for use in writing paper. there were some questions posed on the wiki regarding the notation used in the sequence alignments
- examples of level of detail required: saying that the loop conformation in the intermediate complex may or may not be relevant to the subsequent step in the deacetylation mechanism doesn't allow us to draw a conclusion about how to present this important point in the paper. please provide more details and explain why the literature drew this conclusion. a similar issue arose with the role of the closed loop conformation in the mechanism of Ex-527: there was no definitive conclusion drawn and the status of this task is not clear
- the c pocket binding affinity correlations will be used in the paper for the purpose of showing how we can obtain accurate Kd,NAM estimates and hence Kex estimates based on our models. xg is working on the experimental part of this problem and is finding that the Ki's used in the original regressions are quite different from those coming out of initial rate measurements. you two need to jointly complete these tasks for purpose of paper (including scoring at different levels of theory and running regressions with the appropriate data) soon so we can incorporate this into the paper. this is also related to as's code development on automation of drug discovery workflows
PL(12/2): I will work with XG on literature review and interconnect between experiment and theory.
PL(12/2): I haven't had a chance to look at the draft for second paper. Can you place it under a new folder under Dropbox/PMC-AT Manuscript/?
- as many of the other partially completed tasks as possible from the list should be completed within the next 1 wk so they can start being written up. I assume there has been a lot of time spend on RMSD calculations. At this point, not all custom induced fit development/testing will be directly usable in the paper, but updates on the progress will be useful. If this is taking a long time, we should arrange so other tasks can be completed while progress on IFD is made systematically. If you have any questions about priorities of specific tasks please let me know.
PL(12/2): I agreed that IFD shouldn't be the priority right now as there is no straight usable results for the current paper.
- i will not be able to work further on the paper or provide more information on how the computational results will be incorporated into the paper until closure is brought to more of these tasks. paper writing will be delayed, possibly until 2015, unless we can make more rapid progress on the backlogged tasks
- without more info on partially completed tasks there is little opportunity to provide feedback
Approaches to loop sampling:
- when i said constrained loop sampling options, i meant prime/plop options - did not see any of these yet
- i asked you to comment on whether a distance constraint could be used to force loop building in the appropriate location. please also reply to my question regarding the possible use of plop source code to complete this task.
- please ask promptly about issues where you have questions like setting dihedrals. progress could have been made toward this important task. it is not enough to use the intermediate complex xtal structure since we need to compare energies (see the partially completed tasks where this is mentioned).
PL(12/2): I haven't spent much time on the plop source code simply I do not have time to explore it right now.
I have explored some of the loop refinement approach using prime. And there are options to apply distance constraints. Can you be more specific on what are the target structures and goals we want to achieve?
RC: The goal was to build the loop in the native intermediate loop location starting from 4FVT by constraining it to lie within a certain distance of the native.
PL(12/2): Do you mean we built an intermediate structure (with NAM in C pocket) from 4FVT with loop structure similar to native intermediate? If so, what is the criteria or constraints that you want to choose/make?
RC: Yes, we've been discussing building loop conformation in 4FVT environment that is similar to BVG for some time. I was asking you to consider the possible distance constraints that could be used in this context - that was the task (since I am not looking at the structure, I cannot give exact constraints). The dihedral specification was another approach.
PL(12/2/14): If we are trying to modeling the SIRT3/Intermediate/NAM interaction, it is no better than starting by docking NAM into SIRT3/Intermediate structure. If we are trying to start from ternary structure (4FVT) and investigate the energies associated with loop conformation change, I am not sure changing the loop conformation manually on the ternary structure (4FVT) will provide the information we want. We may be able to prepare the structure you want by exchanging the coordinates of the loops from 4FVT to 4BVG after alignment. Or we may also start with 4BVG and replace intermediate with NAD+ and ac-peptide from 4FVT.
My original exploration of loop refinement is trying to test its ability to predict loop conformational change upon substrate changes, to create an ensemble of loop conformation for IFD, and for fixing the missing loops in Sir2TM.
RC:
Though we are interested in NAM-intermediate complex binding affinity, that is being studied in other calculations on our task list.
We have been discussing the need for scoring the native structure's energy in all our structure prediction and mechanistic studies for some time. For example (earlier postings):
"In each case where structure prediction accuracy is poor we are interested in determining if this was due to scoring function inaccuracy, inadequate sampling, or structural flexibility"
"In any such test, include scoring of the crystallographic structure to determine whether a test failed because of inadequate sampling or an energy function error. "
"Assuming the intermediate complex structure differs significantly from that of NAD+ complex, try to predict the former from the latter using loop prediction, or at least whether the xtal loop structure is one of highly ranked ab initio structures. Energy minimize the xtal loop conformations and compare them to the ranked loop energies to determine effect of sampling efficiency vs energy function accuracy."
Hence robust approaches to such structural substitutions are important for several aspects of our work.
In the present case we are also interested in energy differences for mechanistic analysis purposes (MD simulations are also relevant here).
When we discussed scoring the xtal loop conformation above, substitution of the coordinates is the first thing to consider but a prerequisite for assessing this is the result of the residue-by-residue RMSD calculations from the superposition; e.g.,
"First provide the residue-by-residue RMSD for each residue in loop and also NAM between the SIRT3:peptide:NAD+ and SIRT3:intermediate:NAM xtal structures (see group meeting minutes from ~ 1 month ago)"
since bond lengths and angles may not be chemically reasonable upon direct substitution or other changes in the protein conformation between complexes may not be resolved by simple minimization.
The dihedral substitution approach was proposed (assuming angles could be set) as a robust approach to generating starting structures for specified loop conformations to be used as input to MD simulations and ensemble energy calculations, in case simple methods like coordinate substitution do not work, since it retains appropriate chemical bonding. If the latter works in this case, that's great. (Bonding should be the same in structures compared energetically.)
Constrained loop prediction options were also relevant but the options were not available to me when proposing the dihedral substitution approach.
Constrained loop prediction is also relevant to the other applications you mentioned above so we should keep it on the task list.
We can discuss this further if needed now that the objectives are clear (perhaps at the same time we discuss Arabinda's next development tasks).
Management of AS and cluster:
- You need to take responsibility for as. if the employee does not show progress the direct manager must hold him accountable and provide assistance where needed. i don't think you replied to his comments regarding his inability to meet his recently assigned milestones.
- You need to make sure that issues with the cluster nodes that must be resolved locally do not hamper AS's progress
- We also asked that you look into purchasing of new nodes in this context to speed up md simulations and report on the time required to run these simulations.
- software quotes should be in within a couple of days
- i cannot meet with as to discuss code dev until he completes all the prerequisite testing. this should be done by fri/mon.
PL(12/2): I will work with AS on his assignment, but I do not have enough time to help him diagnose the issues he run into as I am not familiar with the batch server and the protein design code either.
RC: The priority is to make sure that he is ready to start development on our codes, starting with the MM-GBSA code that you know well. We are not ready to plan that since he has not tested it yet.
Regarding the other tasks, he does not update on many tasks - e.g., why did he not finish testing the ftp server yet - and appears to stop working after posting one or two error messages rather than ensuring that other milestones are met. Also, it is not clear whether he understands some feedback given to him. E.g., there may be issues with jobs running on slave001 and I asked him if he had even logged on to slave001 recently - no reply. Then he said the jobs (which may be running on slave001) are queued. Then he said he does not know how to take a node offline from pbs, even though it is described in pbsnodes man page (pbsnodes -o and -c to clear offline status, I believe, but he should check.)
You need to make sure he remains productive.
- i will be only infrequently available after dec 10th due to travel; hence any essential discussions (including giving as new assignments) must be completed prior to that; these discussions will not be possible without further progress on the issues listed above.
PL(11/19): Here is a list of tasks posted before. Some completed, some in progress, some changed.
task_lists.docx
It is hard to put a time on each jobs. Many analyses do not have right tools available and need extra work.
RC: We can start by ordering the tasks in terms of the order of work and check them off when they are complete. Then priorities can be edited if needed. We can also mention the date at which work started on each task.
RC: I would an appreciate an update on this list including the status of the loop sampling (see each of my questions and comments below)and which MD simulations have been set up, so I can provide additional feedback and get the info needed to make decisions on next steps, by Wed.
PL(11/14):
Currently working on:
1) writing script to calculate by-residue RMSD between SIRT3 structures;
Is this the type of scripting that AS could help with based on pseudocode? Please explain why/why not. What language?
PL(11/18): No. The best way for this task is to find the right tools, e.g. VMD (tcl language), BioPython (python), etc, then write a short script to produce result. It will be taking longer time to come up with the pseudocode and teach AS to understand what to do.
2) continue the MD simulation of SIRT3/Intermediate constructed from 4FVT and 4BVG superposition;
PL(11/19): I have found Chimera from UCSF to create movie of MD simulation obtained above. Movie file is located in Dropbox\PMC-AT PLIN\Drug_Discovery\new_SIRT3_Int_traj.mp4. The movie shows 58 ns simulation obtained so far, and there is no sign of converging to the native SIRT3/Intermediate structure, likely due to the energy barrier required to flip the relative position of PHE157 and ARG158 (need to break the salt bridge between ARG158 and phosphate group).
RC: Ok. We will consider alternate sampling approaches after the other tasks are completed.
Is this the PHE we were talking about that occupies the C pocket in the absence of NAM? (I believe we also discussed investigating the dynamics of this PHE in the intermediate complex.)
3) complete the MM-GBSA calculations using the trajectories obtained so far from the above 2) simulation;
RC: Would it take substantially longer to report MM-PBSA energies as well?
PL(11/18): Yes, if it is going to be run sequentially for the whole trajectory. Fortunately, I can break it down into sections and run then concurrently.
4) setting up the MD simulation of SIRT3/Intermediate/NAM from cleaving NAM from ternary structure and C-O bonding formation.
RC:
a)Report backbone torsion angle changes in loop between ternary complex and intermediate complex
Then in intermediate complex models you have prepared starting from the ternary xtal structure (e.g., 2,4 above), set these angles to their values from the intermediate complex xtal structure, followed by side chain optimization and minimization. Report the MM-GBSA/PBSA energies and compare to the low energy loop conformations produced by ab initio loop prediction in prime.
Then set up another MD simulation starting from this structure
PL(11/18): The backbone torsional angles of ternary complex (4FVT) and intermediate complex (4BVG) were analyzed and reported in the attached excel file. The region associate with flexible loop is highlighted (in sheet 1, those with big difference is colored in green). The Ramachandran plots are presented in sheet 2.
By the steps presented above, do you plan to change the backbone of the flexible loop into its structure in 4BVG, then see if the side chain can be optimized? Changing the dihedral angle terms directly can be tricky.
RC: Yes. It would help to know the software options for specifying the dihedrals. This is related to the issue posted on the meetings page regarding the options for constrained loop sampling in prime (note as mentioned we have the plop source and executable as well).
PL(11/19): Not sure how to achieve the dihedral change.
RC: I am assuming prime cannot constrain dihedrals in loop sampling?
If not, what about distance constraints? Constraining distances so they match those of the BVG loop could force loop building with similar dihedrals.
Regarding setting the dihedrals:
--you can first check if any of the protein modeling packages (like amber,charmm) allow setting dihedrals by scripting alone.
--as noted, we have the plop source code: it is in ~rajchak/Simulation/Protein/plop_amit/src. It is in Fortran. This could be used to set dihedrals.
My group has looked closely at the plop source, mapped out various parts of it, and also has been developing a new source code for protein modeling and design in C++, which is located in ~rajchak/Simulation/Protein/protein_cpp_class that has a similar architecture to plop. Such codes can be used build protein structures with specified torsion angles and write associated pdb files.
Documentation is available on the academic wiki.
The torsion angle information is stored in various arrays by plop. (e.g., see the subroutine assphi in build.F as well as the global variables module global_mod.F).
Using a plop input script, it is possible to write pdb files based on all the arrays in memory.
To write the torsion angle arrays would probably require editing some of the write routines in plop source code.
In order to determine the torsion indexing (e.g., what atom numbers are involved in the torsion angles of interest), the current torsion (tor) arrays from either xtal structure can then be written using the input script.
Then, the relevant torsions can be assigned fixed values corresponding to those from BVG through modification of source. The resulting pdb file can be written with the write_pdb input script command.
If you would like to pursue this approach, let me know and I can provide more info.
Are you suggesting calculating the MM-GB/PBSA energies using Schrodinger's suite or AmberTools? Usually MM-GB/PBSA energies are calculated for binding pairs to derive binding affinities.
RC: Plop/prime uses an MM-GBSA energy for the purpose of scoring protein conformations. I am asking for those energies. Which program is used to report them depends on the type of calculation and comparison being made. If you are going to compare to an ensemble average from MD, you may need to do the calculations using AmberTools, since your scripts use AmberTools. You can report plop/prime (opls) values in that case as well. As noted I would like to compare the energy of this loop conformation to those of conformations generated by plop/prime ab initio sampling; this will give insight into whether the issues with the latter predictions were due to energy or sampling errors. Here i am referring not to just one ab initio structure but several low energy structures (including those most similar to the native conformation from 4BVG, even if they were not the most highly ranked). I mentioned MM-PBSA because as we know it can provide more accurate estimates of energy differences.
b) Do simulation 4) for Sir2Tm as well. (If computational resources permit, do a) for Sir2Tm as well; at least the first parts should be done)
PL(11/19): dihedral analysis is performed for Sir2TM (2H4F for ternary, missing residue 37-42; 3D81 for Sir2TM with S-alkylamidate intermediate, missing residue 34-44). Note that a significant portion of the loop is missing.
dihedral_angle_comparison_2H4F_3D81.xlsx
RC: Plop/prime is generally good at building loops around 10 residues in length. Did you try filling in the missing segment? Not clear how the loop conformations compare to the analogous ones in SIRT3.
c) Please indicate whether structures are available to run any of these simulations with Sir2Af2 or SIRT2.
PL(11/19): No. There is not suitable structures to set up these simulations for Sir2Af2 or SIRT2. Docking or superposition may be used to help set up the simulation, but it may be far from native complex structure.
-Please indicate approximate CPU time / ns of simulation for the above MD simulations, when they are started, and also a target date (approximate is fine) for when the results from the non-MD tasks (including 3) will be updated. Based on this info we will decide whether purchase of another GPU node is warranted.-
RC: Given the time required to run the MD simulations starting from 4FVT/2H4F, please provide this info so I can confirm that running the simulation with 2H4F will not saturate our compute resources. (In the latter case, as mentioned, we may consider purchase of a new GPU node for future use.)
d) Meeting with AS asap. All points mentioned on wiki and at group meeting regarding his tasks should be discussed (including next steps w sysadmin including secure ftp server appropriate users should be given access and the folder may be changed if needed). Either AS or PL should post the minutes of the meeting and how the listed points were addressed and AS's schedule for the entire following week.
AS should have reviewed and tested the MM-GBSA scripts within the week.
PL(10/24): The following task list is taken from RC (copied from Workshop, conference and meeting page): And a report related to the tasks below is attached.report_10-24.docx
The IFD reference and supplementary materials are also attached.jm050540c.pdfjm050540csi20051104_093318.pdf.
RC (10/29):
-dataset appears reasonable; need more tabular info on the RMSDs between pairs of structures (see also next point below), indication of which pairs of structures will be compared, and whether structure/ligand pose prediction will be independently tested or the docking protocol will directly be tested as priority
-regarding NAD+/intermediate complexes, if I understand correctly class 1) includes intermediate and 2) includes ternary complex. If so the document appears to indicate a qualitatively different loop conformation between the two. This is related to several other tasks that were posted over last few weeks on group meetings page. Please see those, which pertain to more detailed analysis of these structural differences.
-regarding MD, please start w more info on prior simulations of the the NAM+intermediate complex (from paper 1; see meetings page) as requested vis-a-vis the intermediate/NAD comparison above. next steps for simulations are also provided on the meetings page.
PL(10/30): MD simulations of SIRT3/Intermediate is still running. SIRT3 is taken from ternary complex pdbID: 4FVT, and Intermediate from 4BVG. The approach is better because modify straight from the ternary structure create odd bonding at the beginning and can easily fails the MD run. However, after 12 ns of MD simulation with explicit solvent, the loop structure is still not converged.
Another MD simulation using implicit solvent and GB model is also underway. More details will be reported.
RC: Looking at the ppt from the last group meeting, MD was briefly mentioned in one slide and it wasn't clear which of these simulations were set up anew, which were run previously, and how some of the new simulations were being set up.
Please provide that info.
Regarding the simulation above, does this include NAM? If so, how was NAM prepared? Please see the original comments regarding this simulation. Do you have a MD average from the simulation so far? When was it started?
Also as noted above some more detailed info is desirable regarding the comparison of NAD+/intermediate structures (in absence of MD). Perhaps a closeup of the superposition with comments on which residues are contribution most to the RMSD.
The original plan of work indicated that the prior binding affinity simulations of NAM during PLOS paper could be studied in more detail first to get an idea of the B factors of loop/NAM and how the fluctuations observed during this sampling compare to the distance between the loop and NAM in the intermediate vs the NAD+ complex structure. If more clarification is needed here please let me know. Also please remind me if all our previous MD simulations with NAM had NAD+ in the AB pocket.
PL(10/31): Previously, I have run MD simulations for SIRT3/NAD+/ac-AceS2; SIRT3/NAD+/ac-AceS2/NAM; SIRT3/NAD+/ac-AceS2/iso-NAM; SIRT3/NAD+/ac-AceS2/
1-methyl-NAM; (these three simulations has NAD+ in the AB pocket); SIRT3/NAD+ (all constructed from ternary structure 4FVT).
I have also run simulations with SIRT3/Intermediate; SIRT3/Intermediate/NAM; SIRT3/Intermediate/iso-NAM; SIRT3/Intermediate/1-methyl-NAM (constructed from 4BVG);
I have also run simulations with SIRT3/ac-ADPR and SIRT3/ac-ADPR/Ex-243 (constructed from 4BVH).
Other than the SIRT3 systems, I have also finished MD simulation for Sir2TM/NAD+/ac-p53; Sir2TM/NAD+/ac-p53/NAM; Sir2TM/NAD+/ac-p53/iso-NAM (constructed from 2H4F).
In last group meeting, you have suggested that we construct the intermediate product from the ternary structure by removing the NAM moiety and reconstruct the corresponding bonds. As this will cause some issue for the simulation, my alternative route is to replace the NAD+ and ac-AceS2 in 4FVT by alkylamide intermediate taken from 4BVG.
RC: Looking at the meetings page, I referred to simulation (B) starting after cleavage of NAM (intermediate + NAM). Did you remove NAM from the structure?
The method of intermediate preparation is fine; the question is how you prepared NAM.
As noted if there were questions I would address them were they posed promptly.
PL(10/31): In the current MD, the NAM is removed, because the interest here is to investigate the loop conformation. Since you are interested, I will prepare another simulation with NAM in the C pocket by either superimpose the NAM coordinate from 1YC2 or docked position.
-please let me know if you would like to discuss the above further for clarification
-what about side chain rmsds in dataset?
PL(01/30): the side chain rmsds can not be easily calculated, at least not in Maestro. I will try to find the way to quantify them. By side chain rmsds, do you want to align the complex structures first, them calculate the rmsds with side chain atoms only?
RC: I assume you mean backbone RMSDs (which you provided for loops) are easier to calculate in Maestro.
For sidechains we could either align complex structures or align the backbones of the specific residues. It would be easiest to look at side chains where backbone rmsd is small but the side chain moves, e.g. due to ligand binding.
For purpose of methods validation, i.e. assessment of sidechain prediction accuracy it would perhaps be easiest to start with the problem where we are predicting in the native environment with side chain flexibility.
PL(10/31): OK, I will work out a solution.
-what about SIRT2 structures in dataset? How many are available?
PL(10/30): Currently, there is only 4 structures available (1J8F, 3ZGO, 3ZGV, 4L3O), with two of them (3ZGO and 1J8F, RMSD: 0.44 Angstrom) representing the same apo structures.
-what is total number of structures in Schrodinger IFD dataset? 11 proteins or 11 structures? training set vs validation set? ours?
PL(10/30): As shown in the supporting materials of original IFD paper, there are 11 proteins, (each with at least two structures, some has three) and 23 structures.
I haven't have time to go through all the details of their IDF development paper yet.
RC: In their case, they would only be testing the protocol across pairs of structures of the same protein (hence 2-3 per protein). In our case, since our structures are all of the same protein, I assume we can consider many more pairs of structures
and hence have a relatively large testing/validation set for the protocol despite a smaller total number of structures.
-approach to loop sampling in IFD script. Is constrained loop prediction used? Is unconstrained loop prediction of only certain residues within a specified distance of ligand used?
PL(10/30): In prime loop refinement, no constraint is imposed. The residues within the loop are specified for refinement. A second loop refinement with shorter loop (resid:155-174) doesn't work out well using the default prime loop refinement parameter. We need to re-think about how we are going to modify and test the new protocol.
RC: I meant in the context of IFD not standalone loop prediction. How is it determined which loops to predict in the IFD protocol and does it always specify prediction of all residues in a loop for prediction?
PL(10/31): After checking the input format of IFD, it seems the range of residues for loop prediction is specified manually.
-as noted subsequent priorities for structure prediction/IFD testing will be decided after receiving above info
PL(10/30): The RMSD obtained from Schrodinger of various SIRT3 complex structures vs 4BVG(SIRT3 with intermediate). Classification is noted in the report_10-24.docx.
RC: We should consider which pairs of structures will be used for testing, and for each such pair, what test will be run and the RMSD of the associated substructure (see original workplan)
PL(10/31): As suggested in the report_10-24.docx, I propose to cross dock (using both Glide XP and IFD) not just between pairs, but to dock between the classes of structures as the structure within the same classes are very close and lead to little challenge. And here is some suggested difficult cases:
1) dock ADPR (or ac-ADPR) into 4JSR;
2) dock ELT-11c into 4BVG; (4BVG was chosen because it is a critical structure to our interest.)
and maybe others.
RC: In my reply I indicated that the classes can be used but the detailed enumeration of pairs across classes with indication of what is being tested in each case is important. Please see above and below for the info required before proceeding.
PL(10/31): Here are the list of SIRT3 structures and all these can serve as receptor, but only selected ones has ligands appropriate for study. Still we have many possible combinations for testing ligand docking, and active site structural change if IFD is used. Many docking will likely work well without IFD. I will summarize further other information you requested.
| class 1: |
Receptor |
Ligand |
Note |
| 4BVG |
SIRT3 |
alkylimidate intermediate is not suitible as ligand |
|
| 3GLT |
SIRT3/thio-ac-AceCS2 |
ADPR |
|
| 4BN4 |
SIRT3 |
ADPR |
|
| 4BV3 |
SIRT3 |
NAD+/Ex-527 |
NAD+ and Ex-527 both present |
| 4BVH |
SIRT3 |
ac-ADPR/Ex-527 |
|
| 4BVB |
SIRT3 |
ADPR/Ex-527 |
|
| 4BVE |
SIRT3 |
thioalkylimidate intermediate is not suitible as ligand |
|
| 4BVF |
SIRT3 |
thioalkylimidate intermediate is not suitible as ligand |
|
| class 2: |
|||
| 3GLS |
SIRT3 |
||
| 3GLR |
SIRT3/ac-AceCS2 |
||
| 4BN5 |
SIRT3/NAD+ |
SRT1720 |
|
| 4FVT |
SIRT3/ac-AceCS2 |
carba-NAD+ |
|
| 3GLU |
SIRT3/AceCS2 |
||
| class 3: |
|||
| 4JSR |
SIRT3 |
ELT-11c |
|
| 4JT8 |
SIRT3 |
ELT-28 |
|
| 4JT9 |
SIRT3 |
ELT-3 |
|
| class 4: |
|||
| 4C78 |
SIRT3/AceCS2 |
Bromo-Resveratrol |
|
| 4FZ3 |
SIRT3/ac-p53 |
||
| 4HD8 |
SIRT3/Fluor-de-Lys peptide |
||
| 4C7B |
SIRT3/Fluor-de-Lys peptide |
Bromo-Resveratrol |
| vs 4BVG |
RMSD |
|
| class 1: |
||
| 4BVG |
0 |
Angstrom |
| 3GLT |
0.818 |
Angstrom |
| 4BN4 |
0.835 |
Angstrom |
| 4BV3 |
0.848 |
Angstrom |
| 4BVH |
0.874 |
Angstrom |
| 4BVB |
0.855 |
Angstrom |
| 4BVE |
0.839 |
Angstrom |
| 4BVF |
0.873 |
Angstrom |
| class 2: |
||
| 3GLS |
1.389 |
Angstrom |
| 3GLR |
1.096 |
Angstrom |
| 4BN5 |
1.169 |
Angstrom |
| 4FVT |
0.946 |
Angstrom |
| 3GLU |
1.091 |
Angstrom |
| class 3: |
||
| 4JSR |
1.466 |
Angstrom |
| 4JT8 |
1.537 |
Angstrom |
| 4JT9 |
1.585 |
Angstrom |
| class 4: |
||
| 4C78 |
1.185 |
Angstrom |
| 4FZ3 |
1.092 |
Angstrom |
| 4HD8 |
0.992 |
Angstrom |
| 4C7B |
0.951 |
Angstrom |
| Aldose Reductase |
RMSD |
| 1AH3 |
0 |
| 2ACR |
0.630 Angstrom |
| Antibody DB3 |
|
| 1DBA |
0 |
| 1DBB |
0.503 Angstrom |
| protein CDK2 |
|
| 1BUH |
0 |
| 1DM2 |
1.155 Angstrom |
| 1AQ1 |
1.310 Angstrom |
| COX-2 |
|
| 3PGH |
0 |
| 1CX2 |
1.594 Angstrom |
| Estrogen Receptor |
|
| 1ERR |
0 |
| 3ERT |
0.961 Angstrom |
| Factor Xa |
|
| 1KSN |
0 |
| 1XKA |
0.599 Angstrom |
| HIV-RT |
|
| 1RTH |
0 |
| 1C1C |
0.884 Angstrom |
| neuraminidase |
|
| 1NSC |
0 |
| 1A4Q |
0.217 Angstrom |
| PPAR_gama |
|
| 1FM9 |
0 |
| 2PRG |
0.879 Angstrom |
| thermolysin |
|
| 1KR6 |
0 |
| 1KJO |
0.134 Angstrom |
| thymidine kinase |
|
| 1KIM |
0 |
| 1KI4 |
0.219 Angstrom |
RC: Comments on ppt and results to date on 2). Some comments refer to previous group meeting minutes below.
Sirtuin structure prediction validation:
- Dataset for systematic structure prediction methods development and testing should be provided prior to starting validation. This dataset can be provided on the paper 2 page as discussed. It should include a list of available sirtuin structures (esp SIRT3, but can also include other sirtuins if the SIRT3 dataset is too small), including ligand (drug) complexes wherein loop conformational shifts occur and for selected pairs of such structures, indicate the relevant RMSDs of active site side chains and relevant secondary structural motifs including the flexible loop. These can be used in testing. (Please indicate B factors from xtalography where relevant.) The superposition of structures shown e.g. in the latest group meeting ppt are difficult to interpret visually. See tasks page for more details on how to organize the dataset.
- For each such structure pair, one structure will be used as initial to predict second structure (this includes pairs where the structures are the same and one seeks to predict in the native environment). In each case where structure prediction accuracy is poor we are interested in determining if this was due to scoring function inaccuracy, inadequate sampling, or structural flexibility (energetic degeneracy).
- Eventually, for paper 2, it will be important to do (customized) IFD protocol testing on structurally characterized ligand complexes such as Ex-527 after above are completed to assess false positive rate. E.g., we should be able to show that preferred Ex-527 binding mode is not buried below the C pocket. For such scoring the IFD scoring function may need to be modified as discussed.
Sirtuin structure prediction IFD scoring functions:
- After the dataset above is prepared and while some initial simulations are running, further information on the parameter estimation and testing protocols used to develop and validate induced fit methods in the literature should be summarized. In particular, when did Schrodinger test its advanced IFD protocol and with what dataset.
- A note: We have been planning to develop customized scoring functions for sirtuin structure prediction. In the present context, this can be used to improve structure prediction accuracy on the above testing dataset (need to first assess dataset size and break into training and testing subsamples).
- For later scaling of IFD calculations, note that we have PLOP (older gen prime) source code available and it is used in python protein design script. This can be run on an arbitrary number of nodes. Hence we can do extensive sampling of protein degrees of freedom in our protocols (moreso than ligand degrees of freedom for now) and may later modify IFD to use PLOP not prime as needed. AS may be involved in this.
Conformational changes upon NAM cleavage (please indicate when you plan to schedule these tasks):
- Analysis of past MD simulation data on NAM in C pocket of intermediate complex should be provided as indicated below after our previous group meeting. Please provide analysis of B factors for loop and NAM and a comparison to NAD+ (ternary) complex structure (see below for more details). As noted below MD simulations of the conformational change between these complexes can follow after these results are reviewed and discussed as needed. These are relevant to paper 2.
- Differences in flexible loop between intermediate and ternary complex not yet clear. Is the comparison provided on 4BVG and 3GLS between intermediate and apo, from two different studies? How many intermediate structures are available and from which studies (this may be answered in context of dataset above)? Does the loop conformation differ so substantially in all structures if there are more than one? Please summarize the differences. Please also indicate whether a loop prediction is being run starting from ternary complex structure with NAM cleaved.
- Summaries of MD results, as they become available, should also be organized systematically in a section of wiki page for 2nd paper.
- MD simulation results can also be used to prepare movies suitable for viewing by AS and other group members for purpose of visualizing MD results.
- Please present the results from the loop predictions underway at time of group meeting, as they become available.
- As indicated below results of MD simulations of loops (when they are done) should be compared to results of highest ranked results from ab initio loop prediction to assess adequacy of loop sampling of the long loop (see below for more details).
PL(10/24): The standard procedure of Prime Loop Refinement in Schrodinger doesn't work for the long dynamic binding loop. The refinement of loop from residue 145-181 (37 residues) took 7 full days of CPU time, and the result is way off.
Here is the setting: the SIRT3 and Intermediate are kept fixed as the crystal structure (4BVG) except for the 37-residue loop, the standard multi-step refinement procedure is used. Top ten of the most energetically favorable loop conformations were saved. (OPLS2005 force is used with VSGB solvation model.).
Here is the outcome.
(The crystal structure of 4BVG is in cyan.)
PL(10/20/2014): Proposed procedure for testing of SIRT3 Docking/Induced Fit docking
1) Among 20 SIRT3 Apo/binary/ternary complex (3GLS, 3GLU, 3GLR, 3GLT, 4C78, 4C7B, 4FVT, 4FZ3, 4BV3, 4BVB, 4BVH, 4BVE, 4BVF, 4BVG, 4BN4, 4BN5, 4JSR, 4JT8, 4JT9, 4HD8), identify the binding ligands: including, peptide (AceCS2, ac-AceCS2 (thioacetyl lysine-AceCS2), ADPR, Bromo-Resveratrol, Fluor-De-Lys peptide, NAD+(CarbaNAD), Ex-527, Ac-ADPR, ELT-11c, ELT-28, ELT-3, Thioalkylimidate, SIRT1720, ac-p53-AMC, piceatannol);
2) Test the Glide XP docking of these ligands on the receptor conformations from the above structures;
3) For failed Glide XP docking, run Induced Fit Docking to check the chance of improvement;
4) Modify IFD protocol to change the improvement.
Raj, please common on the above procedure before I proceed further.
RC (10/21):
- the dataset under 1) above should be classified in terms of what conformational changes occur between the structures.
- based on this information, pairs of structures can be assigned to categories for testing specifying whether loop prediction, side chain prediction, ligand pose prediction, etc will be tested for a given pair. This can include protein structure prediction in the presence of the native ligand pose.
- the list can include redocking or loop/side chain prediction in the original protein environment.
- prediction may be done in both directions for a given pair - e.g., if the protein structure changes significantly upon binding a ligand, the enzyme-ligand complex structure may be predicted or the ligand may be removed from the latter structure and the apo structure predicted.
- will provide more feedback after seeing the dataset and its size. other sirtuins can be included if the list is too small. further details of this procedure will be described on the meetings page under the last computational group meeting ppt.
- after seeing the list, we can decide which categories to study first and also decide which complexes should be run through the 10/20 procedure you wrote above (in parallel with the 10/21 procedure). steps 2,3) of the 10/20 procedure check whether changes in side chain and backbone degrees of freedom can be ignored for the purpose of ligand pose prediction in sirtuin-drug complexes. we may choose to include examples where it is not obvious that this is the case. results from the 10/21 procedure will help explain why this is not case when pose prediction accuracy is low. this will be relevant to choosing customized ifd protocols (which relax constraints on various degrees of freedom using various approaches including using ensembles of low energy structures) and will be relevant to publication.
- in the above tests, it will be important to analyze the results in order to determine whether errors are due to scoring function inaccuracy of inadequate sampling.
- in the case that one is predicting structures in the original environment, scoring of native is easy. otherwise, for cross validation, checked sampled structures for the closest one to native to check if the error was due to scoring
function inaccuracy or sampling. (note: in some applications as noted we will use md for this). scripts via AS may ultimately be useful for this
- in this regard please specify flexibility Prime/Glide offer in reporting sampled structures. note the plop executable / source might be used to retain more protein structures from output (see meetings page).
- as time permits while doing the above please summarize validation methods with citations for ifd. which customizations were used in custom ifd reported in lit.
- at customization stage we may estimate parameters for our customized sirtuin scoring function. will advise on what structures to use for training and testing.
PL(10/17/2014):
Task List for week (10/20-10/24):
1) continue to work on slides according to SBIR research plan; (The first draft is available under Dropbox\PMC-AT PLIN\PCR_OPT\SBIR research strategy Phase I.pptx or using the following link:
https://www.dropbox.com/s/6mot5cfmhmxvs44/SBIR%20research%20strategy%20Phase%20I.pptx?dl=0 )
2) help Sherry preparing SF 424 (R&R) and related documents;
3) Make a list of SIRT3 complex structures and schedule a systematic testing case for SIRT3 docking/induced fit docking protocol.
4) Follow up with Arabinda on his progress.
5) Plan on MD simulations of SIRT3/intermediate that constructed from ternary Xtal structure??
6) Set up account for Xiangying & Alok on the cluster, provide the instruction for transferring files to ftp server.
PL(10/14):
/home2 directories on slave003 and slave006 have been cleaned up, and it is OK to remove them if necessary.
RC (10/6):
- when can Schrodinger installation be moved (i.e., when will use of Schrodinger software be idle?) I will also need to unmount slave003 /home from kali at the same time, since as you will
see from the sysadmin nfs tasks, slave003 /home will become the newly shared /home directory. We will then move all your active files on the kali filesystem (including those in /home2) to the newly shared /home directory
- after the nfs to-do tasks are finished by RC, you can a) move all your files from the gpu node /home directory to the newly shared /home directory, and b) backup any old data from your old home directory on slave006 in the /home2 directory on slave003, or if it still being used, consolidate it in the newly shared /home directory on slave003. As you will see in the sysadmin nfs tasks, archived obsolete home directories will now be stored in /home2 on slave003. /home2 will not be actively used.
- later in week discuss cluster administration, starting with new account management. At that point we will start by making a new NIS account for AS
- please describe what if any additional work is needed to complete your mm-gbsa scripts for library screening (including file format conversions if needed). If they are already prepared, we will skip this step and move to the batch server tasks on the sysadmin page later this week.
- AS will be starting on Fri but will realistically not be ready for discussion before next Mon. On Fri, further details may be added as needed to his tasks page.
- after the previous results with NAM-intermediate MD are summarized, a list of validation tests pertaining to the tasks listed on the meetings page should be provided on the appropriate wiki page. We will maintain and update this list with new tests and results until we have a suitable test set for validation of sirtuin structure and binding affinity prediction.
Meet Fri around 1 pm to discuss:
- status of reorganization of home directories; subsequently will remount fileserver and finalize configuration
- software installation user account "app" can be made on brahman.pmc-research.com
- explain to xiangying, alok how to login to ftp.pmc-research.com with winscp and anonymous ftp login.
- review with Wei the labeling tasks for the PMC-AT cluster nodes; discuss additional hard drive space purchasing with Wei
- any updates on comp chem tasks from group mtg
- Initial computational chemistry tasks (esp MM-GBSA scripts and associated batch jobs) for AS (PL to repost MM-GBSA slides prior to meeting).
- Schedule a meeting with AS for Mon 9 am (may ask him to arrive at work at 12 pm).
- Establish schedule for subsequent comp chem and sysadmin meetings ( may hold this meeting every other week in place of regular group mtg; to review cloud development progress, may just ask him to join Th mtg when relevant).
- Setting up Vidyo for meeting (RC will provide AS and SM/AK with vidyo software in advance).
PL(10/07):
-- The Schrodinger installation can be moved anytime.
RC: Are you using Schrodinger software installed on any other machine that requires use of the license server at this time?
PL: Currently no.
-- Please let me know when nfs-to-do tasks are done. The gpu node /home/plin directory files sum up to 412GB. (A lot of them due to MD trajectories.)
-- MM-GBSA scripts is ready for use, but requires extra preparation steps (some may not be easily automated.) It can only run in single thread right now, but we can skip this step for now.
RC: Several sysadmin tasks are completed (see X's on sysadmin page).
I have copied /home/plin from kali to slave003 and made slave003 the new fileserver.
-Please consolidate plin directories on slave003 /home (shared) and /home2.
-/home/plin on slave006 and gpu node are currently not shared, so you can do this. After you are done, I will restart sharing on those nodes.
-Due to the size of the data on the gpu node (which exceeds available free space on fileserver), you can either move most of those files to /home2/plin on the gpu node,
or some other directory, which I can then share across various nodes. The critical files, however, should be moved to /home/plin on slave003 since this will be your new home directory.
-Please also test Schrodinger software in /home/plin and double check that everything else you need is in /home/plin on slave003 at this time, since I may delete the redundant kali home directories thereafter.
-Please also check the Schrodinger subdirectories in /home2/eknoll on slave003 since these are probably obsolete and taking a lot of space. If they are no longer needed, please delete them. There is also Schrodinger software
in /usr/src, probably installed by Eric. Please check these and if obsolete, delete them as well.
-A new user will be added to NIS to manage software. Please confirm that such a user can be used to manage both matlab and schrodinger software. If software is installed in a common directory (see below), we need not install to this user's home directory. Please comment on whether there is any need to have Schrodinger installation files in the managing user's home directory.
-Latest version of Schrodinger software will then be reinstalled, after an update (below) on project tasks, which may require use of the software. You can do this in a couple of places: a) /usr/src (which is shared from slave004, the software dev server) has source code and installation directories of many software packages including matlab.
b) /opt is also available for software installation and can be shared if needed.
RC: We should consider which preparation steps can be automated as tasks for AS, since that is one of his roles.
PL: Some of the modifications required are specific to the protein structure, i.e. changing CYS to CYX for S-S bonded residues, assigning protonation states of HIS. (HID or HIE, or others), special residues in receptor have to assigned manually.
RC: It appears some of these can be automated.
We should start making a list of steps that can be automated related to the initial tasks listed for AS and maintain that list on the wiki (recall we had discussed earlier that such a list should be maintained during your work). Some of this information will be required for further discussion. The previous ppts on the scripts presented at the group meeting may also need to be updated.
An outline of the steps involved in the various drug discovery workflows we have used so far will also be useful.
Please also provide here an update on the tasks from last week listed on the Meetings page. Schrodinger software reinstallation can be done after you have had a chance to use the software to provide an update.
PL(09/18):
Each induced fit docking takes one day to finish. I finished three previous planned IFD tasks (one including extra ligands of Ex-243 and DHP-1b derivative):
DHP-1a,b,c,d: Induced Fit; SIRT3 (from 4BN5): File under Dropbox\PMC-AT PLIN\Schrodinger\DHP_docking\InducedFit_4BN5_DHP1abcd_1-out.maegz
DHP-1a,b,c,d: Induced Fit; SIRT2(from 3ZGO): File under Dropbox\PMC-AT PLIN\Schrodinger\DHP_docking\InducedFit_3ZGO_A_DHP1abcd_1-out.maegz
DHP-1a,b,c,d,b-derivative, Ex-243: Induced Fit; SIRT3:ac-ADPR(from 4BVH): File under Dropbox\PMC-AT PLIN\Schrodinger\DHP_docking\InducedFit_4BVH_aADPR_all_ligs_2-out.maegz
I am currently working on other IFD to other receptors as you suggested earlier, i.e. 4BVG without intermediate. I can begin working on the other tasks you suggested but probably won't have the results new tomorrow.
RC: The only new job mentioned below in 9-17 posting is b); this is a regular docking job (not induced fit). a) was mentioned after last group meeting (see Meetings pg) and c) was on list provided in your group meeting ppt.
Please finish b) as a priority (can you send it 9-19 am?) while the IFD jobs run since it is a regular docking.
If you are waiting for IFD jobs to finish you can, e.g., work on further SBIR ppt edits remaining on list.
Also, in terms of workflow with DHP ligands, unless it is much less convenient with the software, you should send 1c derivative results for each receptor first, followed by completion of other ligands, so we can begin analyzing the results in the meantime.
Since you have already finished c) below, ideally we could have the result of a) with 1c within a day? I am assuming it takes one day to run multiple ligands with induced fit, but less than one for a single ligand.
RC (9/24): Given that IFD of 1c (ethyl ester) with NAM present did not locate the buried binding mode, can you tell by inspection of the earlier buried binding mode results whether the methyl ester or perhaps the acid (which was also tested in the JMC paper) could fit in the C pocket simultaneously with NAM?
For Ex-243 with induced fit, did we find the buried binding mode appearing in any of the results?
PL(09/19): Before starting the docking job, I want to confirm with you the extra setting you are expecting for job a) and b):
a) Do you mean using SIRT3:NAD+:Ac-Peptide as receptor, and place below the C pocket as active site? In this case, it doesn't seem to have enough space to fit DHP-1 derivative.
b) Many of the induced fit docking results have DHP occupying part of the C pocket, I am afraid SIRT3:Intermediate:DHP:NAM can not fit all together. Also, there are many induced fit results, do you want to check out some structure that might still has space open or you want to test only the top ranked pose for each of DHP-1 derivatives?
PL(09/10): Tasks currently underway:
0) Completed the missing loop for SIRT2:ADP structure (3ZGV); Since there is only two residues missing, no homology modeling is needed, loop structures optimized using prime.
RC: Good. You had mentioning building loop based on other SIRT structures. Prime loop prediction is fine (probably preferred since you don't have to align structures). How long is the whole loop? See below regarding loop optimization.
PL(09/11): The whole loop is from TYR139-THR146 that connecting the two domains. There are actually only two residue missing residue PRO140 and GLY141. The b-factors of TYR139 and GLN142 are significantly larger than neighboring residues. The fix is a relatively simple process.
1) Completed docking and MM-GBSA calculation of DHP-1b methyl ester derivative (XG tested) on SIRT3 and SIRT2;
RC: For SIRT2, are these using the structure with missing loop built? If so, what binding mode was identified for SIRT2?
PL(09/11): Both structures (with and without the two residues fixed) were used in the docking.
2) Running Induced fit on DHP-1 derivatives on SIRT2;
3) Analyzing Induced fit results and check if they converge;
RC: I believe you are referring to induced fit on the structure with the missing loop built. As part of this analysis please indicate whether the algorithm is sampling the missing loop conformations with prime. If these do not converge to a buried binding mode, as noted below, it may be useful to prepare a different initial structure and use a modified sampling procedure using e.g. ab initio loop prediction outside of the induced fit protocol. Above you optimized the loop conformation in the absence of ligand. To get a sense of whether this can work, I would need to see the above data and understand the flexibility of this loop.
PL(09/11): What I am referring to here is to check and see if induced fit with different starting structures will lead to similar top-ranking poses, e.g. whether using 4JSR or 4BN5 will give us similar top-ranking binding mode for DHP-1 derivatives.
RC: At the meeting please review the basis for the choice of these particular SIRT3 structures for this study. In the previously posted presentation from last week, alignments were done with 4FVT, 4BVG, (SIRT3) and 4L30, 3ZGO, 3ZGV (SIRT2)
After fixing the loop of 3ZGV with prime, the Glide XP docking results doesn't seem to change much. We still obtained the buried binding mode. The rank order of DHP-1 derivatives remains the same, and the poses are also closely resemble each other with or w/o loop fixing.
RC (9/11): If standard induced fit does not provide the buried binding mode starting with other structures, it will be important to a) carefully assess the differences between those structures and that where the loop was built and the buried binding mode identified; b) analyze which parts of the structures are being sampled by induced fit; c) consider other sampling approaches in case standard induced fit is not adequately sampling the regions of the structures with critical differences. For the group meeting tomorrow, please provide analyses of a,b). Also, please upload the structures obtained so far (including the SIRT2 structure with built loop and docked buried ligand) today to the shared folder.
RC (9/16): Please share induced fit results (from grp meeting ppt list of planned simulations and also RC posting regarding other required induced fit jobs on meetings page) dropbox folder as they become available this week.
RC (9/17): Since we are not having a group meeting this week, please provide the update requested on dropbox and email/wiki today (please share the files you have on dropbox and summarize status/filenames by email or wiki):
The following induced fit / docking calculations should be prioritized:
a) SIRT3:NAD+:peptide - DHP (4FVT)
b) NAM docking to C pocket of SIRT3:intermediate:DHP induced fit result (new)
c) SIRT3:ac-ADPR (from your list of planned IF jobs from your group meeting ppt)
After sharing results in hand today, please try to finish all the above by tomorrow am if possible.
There were other docking tasks listed by RC last week on the group meeting page. For example,
-Compare DHP binding affinities for modes w, w/o intermediate or NAD+ (e.g., 4BVG induced fit calculation reported was in presence of intermediate; compare DHP binding affinities to those in absence of intermediate, where DHP may extend into A pocket).
I assume you saw this and the others posted there.
RC (9-11): Please provide brief replies to the questions above by noon today so we can settle plans for the next group meeting.
Files under Dropbox:PMC-AT PLIN/Schrodinger:
Under folder DHP_docking are the results from MM-GBSA or Induced Fit. MM-GBSA were performed after Glide XP, therefore both Glide XP scores and MM-GBSA results are available in the output files.
The filenames are composed of receptor pdbID; receptor Gird file job#; ligands; Glide XP job#, and prime_mmgbsa-out.maegz
3ZGO is apo SIRT2 structure;
3ZGV is SIRT2:ADP binary structure;
4L3O is SIRT2:Ac-Peptide binary structure;
4FVT is SIRT3:Ac-Peptide:NAD+ ternary structure;
4JSR is SIRT3:ELT inhibitor binary structure;
4BN5 is SIRT3:carba-NAD+:SRT1720 ternary structure;
4BVH is SIRT3:Ac-ADPR:Ex-243 ternary structure;
4BV3 is SIRT3:NAD+:Ex-243 structure;
enz denotes only SIRT3 enzyme is chosen for receptor;
acP denotes Ac-Peptide is also included in receptor;
There are two other in-place MM-GBSA calculation for 4BVH:Ac-ADPR:Ex243 and SIRT3:ELT evaluations.
Under folder SIRT3_vs_SIRT2 includes structure comparison between SIRT3 and SIRT2 (crystal complex vs Glide XP docked structure.) See above for the reference of pdbID.
Under folder SIRT3-Inhibitor De novo Design are the MM-GBSA results on Schrodinger's Core Hopping results based on SIRT3:Ac-ADPR:Ex-243 (4BVH). Top 2500 ligands from the 40,000 Core hopping results were re-evaluated using MM-GBSA method.
PL(08/27): Task list on week 08/25-08/29:
1) Run more induced fit with other receptors (SIRT3:Intermediate, other SIRT3 structures);
2) Summarize results from previous docking/MM-GBSA studies for SIRT3 inhibitors;
3) Plan on screening more SIRT3 inhibitors (using de novo design or available commercial lead-like chemicals);
4) Compare Amber MM-GBSA calculations with Schrodinger's MM-GBSA results;
5) Analyze C pocket of SIRT3 and SIRT2. RC: Please add more details regarding this vis-a-vis my comments on the meetings page and move this task up in priority somewhat.
Specifically the comments regarding the buried binding mode of dhp and
analysis of its overlap with the imidate intermediate. Details provided on meetings page.
PL(08/27): Under Dropbox:PMC-AT PLIN/Schrodinger, folders were created specific to certain projects. And files will be deposited to the appropriate folders.
RC: Please reference the maestro filenames where relevant in the group meeting slides and doc files posted.
Methyl ester tested by XG should also be docked.
Please let me know after homology-based loop building and subsequent standard or induced fit docking is finished. RC (9-9): Please provide a summary of what computations you are currently running in this regard.
As discussed we will then consider possible approaches to sampling the loop and ligand degrees of freedom, including specifying a suitable buried ligand pose as a initial condition for proper sampling.
Under folder DHP_docking, there are output files for DHP derivatives docked into SIRT3(4FVT, 4JSR, 4BN5, 4BVH, 4BV3) using Glide XP/MM-GBSA or Induced Fit Docking, and into SIRT2 (3ZGO, 3ZGV, 4L3O) using Glide XP/MM-GBSA.
Under folder SIRT3-Inhibitor De novo Design, output files from MM-GBSA calculations on top 2000 compounds identified using Schrodinger/Core Hopping (based on Ex-243 and SIRT3:ac-ADPR as receptor) are included.
Files can be viewed in Schrodinger's Maestro by using import structures options.
PL(08/15): More docking results. A quick summary. More Docking results.docx
RC (8/18): Thanks; I assume a more detailed summary of results will be forthcoming. Also, when the Maestro installation is finished, please make the output files available for visualization in a shared folder.
RC (8/6): Please indicate when you estimate completion of the current stage of pcr ppt changes.
Sometime over next two working days (ie this week), please dock compounds 1c and 1b from the pdf posted to the Task List from Lab page (dihydropyridine derivatives) to SIRT3 (intermediate, binary and NAD+/coproduct complexes) and summarize results. Further comments will be provided thereafter.
RC (8/18): Please provide an update on the status of rescoring of leads including those coming out of similarity search as well as the others. Related, please complete the tasks pertaining to the leads listed under 8-7 below.
After this is finished and we are waiting for the rescoring jobs to finish, SIRT2:DHP docking jobs should be set up and started.
PL(08/08): preliminary docking results attached.
Docking of DHP-1cd.docx
RC: Some preliminary points
- Please dock 1b as well.
- Have you run induced fit on the complexes above?
- Please comment on availability of binary (SIRT3:peptide) complexes for docking
- DHP 1c appears to flip in orientation within extended C pocket between the apo and intermediate receptors. Please provide some of the lower ranked poses in each case to assess degeneracy of poses.
- Please provide info on availability of SIRT2 structures for docking
PL(08/12): - Docking for DHP 1b and MM-GBSA calculations are underway.
- I didn't use induced fit on any complexes above.
RC: Please add this to the list of calculations to run.
- Binary structure of SIRT3:ac-peptide is available in 3GLR (SIRT3:peptide in 3GLU). The structure is very close to the ternary structure of SIRT3:ac-peptide:NAD+. The RMSD of 3GLR and 4FVT is 0.6 Angstrom.
I have carried out docking of DHP-1b,1c and 1d to the SIRT3:ac-peptide receptor taken from 4FVT, and the docking score increases suggesting a weaken binding with ac-peptide bound.
- will summarize once all the calculations are done.
- Currently four SIRT2 structures are available. Two of them in its apo form. One is for SIRT2:ADPR, and a new structure become available in February this year is a binary structure of SIRT2:S2iL5, S2iL5 is a macrocyclic peptide with an ac-Lys mimic.
RC (8/7): on 8/6, we discussed several types of information that should be added to the lead lists provided below. This includes scores for the ranked leads, specification of which scoring function was used in each case, etc. Please summarize here the information that we discussed should be added, to verify we are on the same page, and then add it.
Please update on the status of the MM-GBSA rerankings of leads (which I believe have been running).
Also, please provide a summary of the results on leads for intermediate binding in a self-contained format, verifying that the 3-4 slides on this topic from the group meeting talk contain the relevant results. If so, please provide those slides in a separate file so we can easily refer to it when needed.
RC (7/8):
a) what unfinished MOE calculations remain to be done with the new license?
PL(07/22): Current focus is using MOE to convert the obtained results into cvs format, molecular structural format that can be further analyzed by EXCEL, Maestro, etc.
b) Before purchasing compounds, how many off the list can we rerank with MM-GBSA over a two-week period?
PL(07/22): The total scheduled to be re-ranked using MM-GBSA is about 3500 compounds.
c) Did you always use Schrodinger to calculate MM-GBSA?
PL(07/22) For de novo ligand design, the deltaG is calculated using GBVI/WSA dG in MOE. For re-ranking of the available commercial compound, they are re-docked using Glide XP and re-ranked using MM-GBSA in Schrodinger.
d) Regarding loop interactions, did you also find favorable interactions between the loop and the coproduct, such that the closed conformation of the loop prevents coproduct release?
Is the PNAS paper's conclusion about the effect of the loop on coproduct release, mediated by Ex-527, validated?
Did you find favorable loop interactions with all of the top leads? Also for the intermediate binding ligands?
To what extent is the loop conformation changing between structures from the PNAS paper (e.g., intermediate and co-product complexes)?
e) Answers to which of my questions below are still pending? Please provide a schedule of work on these tasks.
f) How would you explain the fact that PLANT converges to solutions that contain larger functional groups with more degrees of freedom, but MOE and Schrodinger do not seem to do so?
g) What other PLANT/Glide correlation tests may be carried out before deciding whether PLANT provides a suitable docking score for our studies?
h) What is the plan for running MD on a subset of leads to check correlations with the other levels of theory?
i) Are the correlations listed in your report lower than those previously obtained for the the C pocket binding ligands?
j) How does N-methyl-NAM rank?
PL(07/03/14): Another report (Part 2) is attached. ShortReport_part2_070314.docx
Some promising leads as SIRT3 inhibitor (against SIRT3/ac-ADPR or SIRT3/NAD+) discovered using Schrodinger (Core Hopping) and MOE (add group) are attached.
Top200_4BVH_3_Glide_CoreHopping.pdf
Top200_MOE_SIRT3_NADp_addgroup2.pdf
Top200_MOE_SIRT3_aADPR_addgroup01.pdf
Top200_MOE_SIRT3_aADPR_addgroup02.pdf
RC (7/2): Please provide an update today on your progress and a summary of topics that will be covered in the report. I am traveling this week and will also not be in the office when you return from your break.
Thus I would like to see the list of topics so that if I have any questions or requests to include some other info I can do so prior to your starting your break.
I assume the report itself will be submitted later this week.
PL(07/02/14): The report will cover some answers to your questions/requests post below. And the majority of the report cover the exploration of the MOE software. The various tries and new leads identified using MOE are included. Also, new LEA3D calculation using the complete library provided. Also in progress are the identifications of new testing molecules for experiments from the leads (excluding those may lead to patent infringement.)
CCG also contacted me offering to extend the trail period of MOE. I think we should take advantage of that.
PL(06/23/14): Still working on the report and schedule.
PL(06/24/14): Short report Part1: ShortReport_part1_062414.docx
RC (6-25): Comments/questions on the short report and next steps:
a) time estimates for calculations at various levels of theory:
-vis-a-vis the plan for programmer tasks accelerating throughput, how are you running MM-GBSA calculations - on one node?
We will consider plans for implementation on batch server.
-please list any other potentially "automatable" steps that are proving to be bottlenecks in the work to date
PL: The MM-GBSA calculations is carried out by Schrodinger and is limited by the license. It can only run with single thread.
Same situation for Glide docking. The license limitation is the bottleneck right now.
b) growing using MOE:
-is it being done using NAM as starting scaffold?
-since MOE license will expire within a week please post schedule for remaining work with it asap
PL: currently, the add group approaches all start with amide group, with SIRT3/ac-ADPR and SIRT/NAD+ as receptors, which provide a larger selections of scaffolds, which include six member rings connected directly to amide group.
NAM is docked to the SIRT3/ac-ADPR, but add groups to NAM is not started yet.
c) correlations between binding affinity estimates at various levels of theory:
-please provide this for a subset of compounds from the list that have been or shortly will be ranked using various levels of theory including MM-GBSA.
PL: Below graph shows some results of binding affinity estimates at various levels of theory:
Among top 50 ligands (39 unique) generated from Glide_based core hopping against SIRT3/ac-ADPR receptor, Glide docking is performed using various level of theory. MM-GBSA is not performed because the license is still in use.
39 top ranked unique ligands and their scores were plotted below.
(docking_score_CoreHopping is the scores directly from CoreHopping; HTVS/SP/XP score is the Glide score after re-docking the pose at HTVS/SP/XP levels)
SP results correlated best with the scores from CoreHopping (at HTVS level). However, in Core Hopping, constraints is applied to the amide group and Chlorine atom. With XP docking results changes significantly.
d) ranking:
-of NAM,isoNAM, N-methyl NAM against intermediate binding ligands - this may be more informative than ranking against the Ex-527 mimics.
The lead discovery using SIRT3/Intermediate as receptor is not extensively explored before license expiration. One set is obtained using isosteric matching with Ex-527 as template (keeping the amide group), and the 15774 leads are not unique for SIRT3/Intermediate. The docking of this set against SIRT3/Intermediate using HTVS was completed before and the best docking score is -9.428, and best docking score for NAM/isoNAM is -6.416/-6.739, which ranks below 6651 among all 52470 poses obtained using 15774 leads.
Another set of lead was obtained by core-hopping using the modified Ex-527 as template, 346 leads were obtained using the small linker library. The best HTVS score is -8.416, and isoNAM rank 34 among the 346 leads.
e) LEA3D:
- what tests are being run with LEA3D (what previously determined leads, if any, are being "optimized" with it)?
- noting that Ex-527 ranked # 42 (higher than mimics), can we test whether LEA3D can convert mimics to Ex-527, or is it outside of scope of LEA3D?
- vis-a-vis programmer tasks, could it also be run in batch mode, or could a single optimization be run in parallel on multiple processors?
PL: The test on LEA3D so far is very limited. The sample jobs were all tested.
The current test job with LEA3D is not a true production test, because the library used are not fit for real lead optimization. The test currently running is for the purpose of learning the setup and time requirement for real application.
LEA3D can not be run in parallel. One possibility of speeding up is to set up multiple jobs from multiple leads, and run it at the same time.
Currently, I am using the PLANT code for docking, I am trying to see if we can use it to re-rank the output from Schrodinger and MOE, and it will be great if there is a good correlation.
f) is residue interaction analysis between Ex-527/loop (vis-a-vis mechanism discussed in PNAS paper) and Ex-527/NAD+ to be scheduled in plan?
The raw data is obtained. I will start the detailed analysis after the MOE license expired.
PL(06/06/14): Weekly schedule (06/09/14-06/13/14):
1) Analyze completed tasks.
2) Continue MOE test runs with different targets(receptors). Try other tools in MOE's fragment based design and RECAP analysis.
3) Start setting up LEA3D calculations.
4) Compiled some promising hits for experimental validations.
More tasks will be scheduled once the above tasks are completed.
PL(06/10/14): Further breakdown of tasks:
1) Determine and create workflow to discover leads with selectivity for SIRT3/Intermediate (vs SIRT3/NAD+ and SIRT3/ac-ADPR);
2) Try MOE to find new leads (one option is to grow from NAM);
3) Try LEA3D to set up optimization targeting specificity of the lead (to certain target);
4) Plan on the programming requirements for automatic workflows;
5) Make estimates on the time required for compound docking (HTVS, SP, XP and MM-GBSA);
PL(06/06/14): Work performed after group meeting.
1) Calculate glide score for Ex-527, NAM, isoNAM using Glide at HTVS level of accuracy, using SIRT3/ac-ADPR as receptor (same as the one use for screening.)
2) Set up MM-GBSA calculation using the leads obtained using glide-based core hopping. (No re-docking at higher accuracy is performed.) Job currently running.
RC: Post correlation between glide sp, glide xp, and mm-gbsa for a short list of molecules and then also correlation between mm-gbsa and md and/or induced fit for a subset of mm-gbsa highest ranking molecules. Also
post total time required to obtain all data points for each level of accuracy.
3) Set up MOE to run scaffold hopping using larger linker library (762K vs. 20K in the original test)
RC (6/3/14): Please post the minutes for your next tasks from the group meeting here, including action items proposed by all attendees.
These include:
a) ranking Ex-527, NAM and isoNAM among the hits
b) further residue interaction analysis, including but not limited to investigation of the role of the closed loop
c) reranking the hits based on higher level of theory - including MM-GBSA. I am curious to see the extent to which the rankings change.
Time estimates for scoring a given library size with the different levels of theory.
d) continuing studies with the MOE trial license (including intermediate complex). An important question is, how would these licenses be used going forward for a full year, if we are able to generate hits using large core libraries within a few days.
e) applying LEA3D to further evolve hits from Schrodinger or MOE (a type of automated lead optimization)
We identified an important difference between core hopping and LEA3D in the approach to optimization. Can LEA3D run on multiple processors?
f) similarity search of hits against commercial databases, followed by ranking of the similar compounds, for the purpose of experimental validation
g) checking the binding affinity of the intermediate complex hits to other complexes (NAD+, coproduct complexes) - this is important, since we want to determine whether there is scope for -selective- binding to the intermediate. Residue interaction analysis may help.
h) trying other halides as one side chain
i) discussion of next steps with Schrodinger license: building from NAM/isosteric matching?
j) providing more details on how LEA3D genetic algorithm operators work
RC: I look forward to the schedule of planned work for the next week including any items from the meeting that I missed above.
PL(05/27/14): Proposed outline for group meeting: computational part
RC (5-27): Regarding topic 1, note that Steegborn (PNAS) mentioned that Ex-527 stabilizes the "closed" conformation of the active site, preventing (co)product release. He may have referred to favorable interactions between Ex-527 and the flexible loop. Will this hypothesis be analyzed from the perspective of MD simulations?
Regarding topic 2, please mention somewhere the MOE drug "rediscovery" test we discussed so we have a point of comparison for what we hope to eventually achieve with the Ex-527 tests.
Please have one backup slide on the computational results with N-methyl NAM (not a central topic of presentation).
Please post the draft sections of the ppt as they become available (not necessarily final versions).
PL(05/28/14): I will set up calculations to analyze the residue-ligand interactions to see if any support for this hypothesis can be found.
I am working on MOE with demo license, and will show results once available and will modify the outline accordingly.
RC: Ok, if the results are not available it is fine for the purposes of the group meeting to summarize the example given in the MOE paper.
PL(05/9/14): Weekly schedule (05/19/14-05/23/14):
1) Discuss with Schrodinger about the small molecule discovery suite, especially the de novo design using Core Hopping, etc.
2) Prepare long Schrodinger report.
3) Test run using LEA3D. The following three references are about the LEA3D method. The programs are mainly written in Perl, while calling many open source/free/proprietary software for processing input files, file conversions, docking, pose rescoring, property predictions. It may be time consuming to get it work for us, but we may build our in-house codes using the same concepts.
Nucl. Acids Res.-2010-Douguet-W615-21.pdfjm0492296.pdfart%3A10.1023%2FA%3A1008108423895.pdf
RC (5-24): You a mentioned long schrodinger report - will this include corefinder review? What is the status of the work with corefinder to set up the Ex-527 rediscovery? Do we need to request a new license to do any more work with this?
Will lea3d be used for rediscovering ex-527?
Given status of the above, which topics will be covered in group meeting, if the meeting is held within the next week or so? Please augment lea3d slides with schrodinger/corefinder and example of ex-527 applications; perhaps more discussion of the moe rediscovery example so we can compare our plans to it; also, ex-527 md results and analysis. Based on the progress with this we will finalize the group meeting date.
RC (5-27): Once I receive the outline of the computational ppt for group meeting today, we will decide whether to hold the group meeting on Fri of this week or Mon of next week and what the agenda of the meeting should be. If we conclude there is enough to concretely discuss the plan of work based on the progress to date, it is ok to hold the meeting earlier.
If it is helpful we can decide precise content after I receive the outline.
PL(05/27/14): I have tried corefinder, which is able to extract core patterns from given input structures. It doesn't require extra license to run. By using corefinder, I was able to look into the structures that forms the database used for core hopping in Schrodinger. It seems there is a mechanism in core hopping to merge different rings, but there is no information currently available on how they did it.
Using Ex-527 as input structure, corefinder is not able to create any core library. (reason unknown). I do plan to request a new license to explore further on the application of schrodinger's small molecule discovery suite.
I have set up LEA3D to do anything beyond the examples. But it is possible.
I will present on the group meeting about the drug design based on Ex-527. I plan to talk about what Core Hopping is able to do for us so far, given as a black box. I also plan to talk about LEA3D as a drug design program, the workflow, and its potential for our drug discovery goal.
I recently requested the MOE download and demo license and if possible, will try to run some tests using it.
I will also present Ex-527 MD results and analysis on the group meeting, which is considered complete.
The presentation preparation will be time consuming. Other than what i prepared before, I plan to add more background information, testing, result and analysis section to the presentation. I will try to provide an outline once I decide on how to organize all the materials.
PL(05/20/14):
After talk with Thijs from Schrodinger, here is what I learned. 1) Thijs mentioned that the core hopping module can do routine de novo drug design by combining fragments, core and linker. There is no specially treatment as to ring fusion or cyclization. They will be happy to supply the detailed implementation method if we are going to publish the results. The core library is the main source for new scaffolds. 2) He suggested an utility tool can corefinder to generate new core fragments, which we can use to "re-discover" Ex-527. 3) The synthesizability score is not a real retrosynthesis analysis, and has little significance.
The plan for next step Schrodinger test: 1) Use corefinder to build new core library including Ex-527, run core hopping using modified Ex-527 to make sure Schrodinger can "re-discover" it with high ranking. 2) run new tests with largest core library to find new hits for SIRT3 with AADPR (the product) and SIRT3 with intermediate. Test the efficiency of the programs.
RC (5-20): This plan sounds good. We can discuss the later steps further at the group meeting if the early steps are still in progress. Some further details on corefinder would be useful. Right now it sounds like Schrodinger recommends it as a black box and will provide details of implementation only later closer to publication time?
Plan for LEA3D: 1) Understand the workflow of the program; 2) Run sample tests; 3) Make use of the programs to generate new leads. Test the efficiency of the code. Currently, the code appears to be somewhat slow in structure-based design even with relatively small fragment library.
RC: Once 1) is done please provide details here comparing to MOE (and Schrodinger) to whatever extent possible.
PL(05/23/14): Now I have run all the examples shipped with LEA3D excluding those require licensed software. Attached is brief summary of the drug design approach provided by MOE and LEA3D. LEA3D is quite flexible and can be modified for new implementations, but are tedious and difficult to run, and it can be really slow for generating and sampling large fragment database.
LEA3D_summary.pptx
PL(05/12/14): Weekly schedule (05/12/14-05/16/14):
1) Complete the data analysis on results from Schrodinger trial license.
2) Send email to Schrodinger requesting details/documentation on the workflow and implementation of Core Hopping module, mentioning related scaffold replacement in MOE.
3) Attending Schrodinger User meeting at Princeton on Thursday (May 15th).
4) Study the LEA3D code and find out if it is suitable for our drug discovery. ( I have downloaded some extra program required to run LEA3D).
PL(05/09/14): Here is a brief report on what has been done using Schrodinger's suite of software in the Drug discovery based on Ex-527. Brief report on Ex-527 based drug design using Schrodinger.docx
Further analysis and report will continue next week.
I will also get in touch with Schrodinger scientist regarding the questions we have on the software next week.
RC: Please let me know a) once you have organized a list of questions for the Schrodinger scientist (vis-a-vis completion of data analysis from the trial license if required); please include here reference to
MOE functionality as we discussed. b) your plan for testing the other open source scaffold hopping software we recently received.
Based on this schedule I will decide when to assign you the next steps for the PCR presentation, which will be to combine a ppt we have prepared on our own technology with the rapid cycle PCR ppt to make an industry demo
ppt or teaser that can be used in discussions with companies.
I will be attending the Schrodinger User meeting at Princeton next Thursday (May 15th).
RC: Regarding the compound similar to Ex-527 in step 3, was there any Glide analysis done?
Which compounds were generated using Glide-bases screening?
PL(05/12/14): the structure-based core hopping in 3) & 4) on the report, is Glide-based ligand design. A receptor grid file is provided for screening in the process. No further Glide docking was performed on this class of ligands.
PL(05/08/14): Weekly schedule (05/05/14-05/09/14):
1) Help finish PLOS ONE manuscript submission;
2) MD/MM-PB(GB)SA calculations for Ex-527 in SIRT3:NAD+ and SIRT3:AADPR are now completed and report will be generated by 05/09/14;
3) Complete the testing tasks for Schrodinger's drug design suite, analyze results and prepare a report.
PL(05/09/14): Results of MD/MM-PB(GB)SA calculations for Ex-527 in SIRT3:NAD+ and SIRT3:AADPR are presented in the EXCEL file attached.
SIRT3_Ex243_MM-PBGBSA.xlsx
The reported Mean values of binding affinities are averaged over last 16 ns from a total of 23.6 ns of MD simulation. The component analysis (on second tab) is averaged over 22 ns.
The result seems to suggest that Ex-527 may actually enhance the binding affinity of AADPR instead of provide a strong binding itself.Structural analysis can be carried out if needed.
The two MD simulation runs suggested that SIRT3:NAD+:Ex-527 and SIRT3:AADPR:Ex-527 are both rather stable with average RMSD at about 1.3 angstrom.
RC: To understand whether AADPR binding is enhanced, wouldn't we need to compare AADPR affinity to the complex without Ex-527?
PL(05/09/14): Yes, if we are going to derive the conclusion from the computational point of view. The suggestion above is based on the computational results that Ex-527 binding affinity didn't change much with AADPR vs NAD+, and binding affinities of AADPR is larger than NAD+, together with the experimental observation for the PNAS paper that Ex-527 together with AADPR is able to inhibit the reaction, while Ex-527 and NAD+ is not able to. I would suggest we carry out the binary system simulation of SIRT3:AADPR to test the hypothesis if that is of our interest.
RC: Ok.
PL(04/28/14): Weekly schedule (04/28/14-05/02/14):
1) Try Core Hoping, and other drug design tools in Schrodinger suite;
RC: Please provide an update on this and the use of BRICS, LEAD3D and/or GANDI. I forwarded the response from at least one open source provider.
PL(05/01/14): BRICS is the one we got the source. It is a database with a collection of molecules in mol2 format and linkage rules written in FTrees can understand. It is of little use to us for now.
I have tried out ligand-based core hoping that can easily generated thousands of new ligands using supplied core structure database. However, there are limitations using Ex-243 as template. The R-groups can only be amide and Cl, which reduce the many possibilities. I am also playing with glide-based core hoping, and haven't got much success so far. There are jobs that are running. I will play with other features once these are done.
I will also look into other open source software after done playing with Schrodinger suite, such as LigBuilderV2, AutoClickChem, and AutoGrow 3 (source obtained).
RC (5/02): Since there is limited time remaining in the schrodinger Core Hoping license, and I may like to consider some tests to run with it, please provide a summary of its features including what types of cuts are possible and whether the possible
cores are all listed in the database or generated through combinations of moieties in the database. As discussed we would like to try to rediscover Ex-527 with core/scaffold replacement tools.
Also, if you have any information regarding the history of Ex-527 development in terms of the lead compounds that were eventually optimized to Ex-527, it may be useful in doing the above study. We could then
start with the lead compounds and try to hop to Ex-527. Note the MOE test case did something similar - starting with a lead compound discovered experimentally and then carrying out lead optimization to see if the clinical candidate could be generated computationally.
Finally, please send me your draft emails for LEAD3D and GANDI and I will send them.
PL(05/02/14): After playing with Schrodinger's Core Hopping module, and reading its document, fragment database files, linkers description file, it appears to me the what Core Hopping is doing is to preserve at least two R-groups (end groups) and explore the core fragment database (file in SQLite format, fragments can not be visualized in Schrodinger, and I can locate some SMILES strings in the file) and apply the linkage rule (which includes 1-atom up to 4-atom substitutions, but I am not sure how they are implementing the rule.) From Ex527, there is little choice for R-group, which has to include heavy atom and not in a ring structure.
RC: Regarding the latter limitation, based on our discussion of MOE (which mentioned the ability to sample rings), do you feel that Schrodinger's core hopping is more limited than some of the other available software? Please let me know.
PL(05/08/14): Today is the last day for the trail license. There are tools in Schrodinger can do some interesting things, such as keeping the amino group and explore structures similar to Ex-527 by exploring the core library. I haven't figured out if Schrodinger can do ring fusion or other features in MOE. From the large number of structures generated using Schrodinger, it appears it might, but I will have to check the results and/or consult with Schrodinger's help desk. I will report it later.
RC: How about sending an email to Weston today or tomorrow indicating this question and how you would like to explore this feature with the trial license, but have not yet been able to find out how to do it? (vis-a-vis mentioning MOE software can do it)
RC: Please let me know when you expect to prepare the report on Schrodinger drug discovery tools.
Without knowing what is included in the core fragment library database, I set up new test using the following template structure.
The R-groups were defined as Cl- and NH2C(=O)- . I have chosen a larger database and the job is currently running. With the new template, I will try to see if we can keep more features for core hopping.
The Ex-527 is obtained through high-throughput screening experimentally using Fluor de Lys assay. (first reported on Soc. Biomol. Screen. 10th Annual meeting, poster P10045, 2004.) The paper published on 2005 on J. Med. Chem. (attached.) jm050522v.pdf And there are two follow-up papers in 2006. (J.M. Solomon, R. Pasupuleti, L. Xu, T. McDonagh, R. Curtis, P.S. DiStefano, L.J. Huber, Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage, Mol. Cell. Biol. 26 (2006) 28–38. | V.M. Nayagam, X. Wang, Y.C. Tan, A. Poulsen, K.C. Goh, T. Ng, H. Wang, H.Y. Song, B. Ni, M. Entzeroth, W. Stunkel, SIRT1 modulating compounds from highthroughput screening as anti-inflammatory and insulin-sensitizing agents, J. Biomol. Screen. 11 (2006) 959–967.)
Its discovery is not by computer-aid drug discovery, instead, Ex-527 is first identified and efforts were put to improve the potency. Some useful discussion can be found in the J. Med. Chem paper attached.
RC (5-8): Ping, can you provide an update on your work on scaffold hopping, in particular with Schrodinger tools since that license is expiring?
RC: Is this paper cited in the J Med Chem paper available?
Hixon, J.; McDonagh, T.; Curtis, R.; Distefano, P. S.; Napper, A.; Hesterkamp, T.; Thomas, R.; Keavey, K.; Pons, J. Discovery of potent, selective, orally bioavailable inhibitors of the human deacetylase SIRT1. Society for Biomolecular Screening, 10th Annual Meeting, Orlando, Florida, 2004.
PL(05/02/14): No. I did not find the poster. Instead, this paper may include something in the poster. Mol. Cell. Biol.-2006-Solomon-28-38.pdf
And this paper followed the same protocol and also reported Ex527 in a later time. J Biomol Screen-2006-Nayagam-959-67.pdf
2) Continue MD with Ex-243 and NAD+/AADPR;
PL(04/18/14): Weekly schedule (04/21/14-04/25/14):
1) Get source code of BRICS, LEA3D, GANDI;
2) Find recent reviews on De novo drug design;
3) Contact Schrodinger about Core Hoping and drug design modules;
4) Set up MD for Ex-243 with NAD+ and AADPR;
5) With free software, explore their capability on scaffold-hoping and drug design;
PL(04/14/14): Weekly schedule (04/14/14-04/18/14):
1) Report on scripts for MM-PB(GB)SA calculation & residue interaction analysis;
2) provide information on relevant pharmacophore and drug-like molecule generation software and procedure;
3) test on computational Kd - experimental Ki correlations with project 1 C pocket binding small molecules for various lengths of MD simulation
4) Begin preparation for Ex243 MD simulation and binding affinity calculations.
PL(04/15/14): Report of scripts for MM-GBSA calculation & residue interaction analysis: MM-GBSA calculations for poses.pptxMM-residue_analysis_code.pptx
PL(04/09/14): Weekly schedule (04/07/14-04/11/14):
1) Report on PCR survey; (done)
2) Structure analysis related to Ex-243 and docking results; prepare a report.
PL(04/11/14): Report on Ex-243 structural and binding analysis. SIRT3 interactions with Ex243.docx
RC: Questions regarding the report:
a) What is being shown/compared in "only small structural changes" fig
b) Need residue interaction analysis including that for NAD+/product
c) Why are the product/intermediate complex residue interaction diagrams not shown
d) What was the Ex-527 rmsd of the (docked) AADPR complex compared to xtal - i.e. can we predict the pose of the drug using other crystal structures,
due to the only small structural changes mentioned?
e) When our GPUs are idle, a next step on this project would be the task below on setting up MD binding affinity simulations for these complexes.
PL(04/15/14): Answers:
a) The small change refer to the two structures above (4BV3{SIRT3/NAD+/Ex-243 complex} and 4BVH{SIRT3/2’-O-acetyl-ADPR/Ex-243 complex}). The RMSD for two protein is 0.677 Angstrom, the RMSD for Ex-243 after protein alignment is 0.438 Angstrom.
b,c) The analyses for NAD+ and AADRP are now included in the updated report. (with the same file name: SIRT3 interactions with Ex243.docx). note that there is a clash between ARG158 and NAD+, probably due to the X-ray crystal analysis issue.
c) Since the focus is on Ex-243 in the C pocket, other interaction diagrams are omitted.
d) RMSD of Ex0527 between top docked pose and Xtal using 4BVH as receptor is 0.136 Angstrom. The small changes only refer to difference between 4BV3 and 4BVH, and changes vary for other structures (i.e apo-enzyme, other inhibitor bound structures.) Using other structures, the top pose may change, and the binding affinity may change as well.
e) I will get it started after I finished some requested works.
RC: Please provide some estimated completion dates on the tasks below. In particular, please post some information on the scripts you have written for MM-GB(PB)SA calculations and residue interaction analysis by early next week.
Please also at least provide links to product feature info on the relevant pharmacophore software and library search software mentioned below by Mon.
moe_scaffold.pdf
RC: I have reviewed the attached and considered plans for implementation of such strategies in our group. Please make sure to read it as well.
PL(01/14/14): There are more information available on fragment-based drug design on CCG's website. https://www.chemcomp.com/MOE-Fragment_Based_Design.htm
However, during the last conversation with CCG, the MOE provide complete package for $50K/year. I have been searching for open-source alternatives. One of them is crystaldock (http://nbcr.ucsd.edu/data/sw/hosted/crystaldock/). e-LEA3D from http://chemoinfo.ipmc.cnrs.fr/lea.html seems to offer some tools for our needs but only free to academics. Please let me know what you have in mind for implementation. Let me know when is a good time if a meeting is necessary.
RC: Can these be run from the command line? I only saw a web server for e-LEAD3D.
We will aim to have a discussion on the plan soon, after I receive some of the info above (e.g. current scripts being used in our group).
RC: One of the bottlenecks in the development of our own tools (based on open source software) for lead optimization is the scaffold hopping/replacement step. Many of the other steps that are part of the MOE workflow are not required
and the cost is therefore not necessarily justified. (Also, these algorithms do not rely on proprietary scoring functions like Glide, which can command a higher price.)
We should carry out an analysis of the algorithms for scaffold hopping/replacement that are in the published literature in order to better understand how difficult they would be to implement.An example is Recor:
http://www.ncbi.nlm.nih.gov/pubmed/19671127
Please download these publications and post here, toward the end of developing a pseudocode for some of the available algorithms.
Also, please confirm what command line open source alternatives are available for this step. You may also check whether any trial licenses are available. If pricing is available for other commercial tools, that info will also be useful.
I will also check with colleagues who may have access to tools for scaffold replacement that do not require licenses.
It may be computationally more tractable to filter hits based on synthetic viability after optimization rather than during optimization. This is also easier to code, since the filtering need not necessarily be done from the command line. There are several retrosynthetic analysis algorithms for which software implementations may be available. Please look into this as well.
We will then have a meeting to discuss possible approaches to implementation of a lead optimization code that integrates scaffold replacement with other steps including binding affinity optimization, leveraging our software under development for protein design - which is similar but replaces the scaffold hopping step with mutations, adding new atoms and building new bonds based on template files for the amino acid residues rather than ligand libraries.
PL(04/17/14): The correct link to the ReCore reference is http://www.ncbi.nlm.nih.gov/pubmed/17305328. The article titled "Recore: a fast and versatile method for scaffold hopping based on small molecule crystal structure conformations" is attached. ci060094h.pdf
I have looking into available resource and approaches for ligand design, and here is a collection of what I found. Report on Ligand design software.docx They are categorized with short description.
RC: We will need to look carefully at the open source options from this list. A more detailed list of features of each of those would be desirable along with references in dropbox. How about retrosynthetic analysis to filter for synthetic viability - do all these programs do that as well?
PL(04/18/14): I am reading some of the references on the ligand design methods. Some of them are coming from Prof. Matthias Rarey's group at The Center for Bioinformatics (ZBH) of the University of Hamburg. Some of their methods are now commercialized, such as ReCore, FTrees, etc. It seems the retrosynthetic analysis is often inherently included in those codes in the selection of linkers of fragments, or the cleavage rules to generate fragments and linkers, or the reaction rules that used to create new molecules.
References for all the codes in the report and details description may take a while to get. I can work on it next week if you want me to. Or we can choose some most likely path/software to look into the details. I have downloaded some of the codes, as open-source or only executable.
RC: If time permits I will stop by today to discuss the overview of the available open source codes (which are most likely) and the retrosynthetic analysis issue. I seem to recall reading in MOE that they appear to filter the hits based on retrosynthetic analysis after running the replacement algorithm.
PL(04/18/14): Indeed, the MOE report did discuss about retrosynthetic analysis at one point. This retrosynthetic analysis usually depends on the starting materials database and reaction rules. One of the method mentioned BRICS is available for free use but requires an institutional email address.
I will be in the office until 5.
And here I posted a few references from my collection, if necessary, I can keep a copy in dropbox. ci900287p.pdffebdey08.pdfjournal.pcbi.1002380.pdf
It will be a major project by itself to develop a proprietary ligand design code. We may be better start with some open-source and/or cost-effective commercial options, so we can focus our effort on drug discovery.
PL(04/08/14): Report on Primer design & related software survey. Primer Design Software.pptx
RC: The fast PCR survey may be augmented with our own PCR control slides shortly for future use.
RC (4/08/14):
- Ping should provide docking/single structure binding affinity calculations of Ex-243 – for both intermediate and product complexes - and also a summary of available crystal structures for computational analysis
- Ping should list the important contacts made by Ex-243, in particular describing the pocket into which the aromatic rings project. He should verify that Ex-243 binds weakly to intermediate due to the steric clash.
- Ping should provide more information on the drug library generation protocol based on specification of a motif (pharmacophore), with details of how he chose the ones used so far.
- Ping should provide more info on MOE pharmacophore software including the details of the mutagenesis algorithms available and how they can be called from the command line.
- Ping should describe functionality of the scripts he has written for MM-GB(PB)SA calculations and residue interaction analysis.
- Do benchmark studies on computational Kd - experimental Ki correlations with project 1 C pocket binding small molecules for various lengths of MD simulation
- Carry out MD binding affinity calculations on Ex-243 intermediate and product complexes.
- Carry out residue interaction analysis for Ex-243 intermediate and product complexes.
- Estimated completion dates for the tasks above should be provided when possible. A group meeting will be scheduled to review results from some of the above tasks, as well as those from experiments related to this project.
- SIRT1 structure preparation will be discussed later.
Upcoming tasks:
- Updating of cluster NFS implementation
- Updating of cluster batch server implementation
- Discussion of drug discovery software development vis-a-vis protein design python/C++ code previously developed by RC group. Such projects will be pursued in order to automate the PMC-AT mechanism-based drug discovery workflow.
- Consideration of add'l hardware needs if any
PL(04/04/14): Report of fast PCR survey. Fast PCR Survey v2.pptx
The survey doesn't include the software for Primer design, which I will put on another report.
RC: I have reviewed the above and we can discuss when we meet. Is the primer design report the first task for this week?
PL(04/07/14): Yes. The task I have right now is to put together the information I collected for the prime design. And I am looking forward to the meeting so I can lay out the tasks for the rest of the week.
RC: Please let me know when you plan to submit the primer design report since we may discuss it at the meeting as well.
PL(04/07/14): The report should be available by the end of tomorrow.
RC: Regarding the residue contribution analysis for sirtuins, is there a summary of results available?
PL(04/07/`4): I have carried out analysis for some systems and some of the results are put into Excel file, but no further analysis and summary are made so far.
PL(03/21/14): weekly schedule (03/24/14-03/28/14):
Task lists based on priority:
1) Structural comparison of C pocket binding residues (alignment & RMSD) in the form of NAM moiety of NAD+ or NAM standalone between Sir2Af2, Sir2TM, SIRT3, SIRT1, in both apo form or with ADPR/NAD+ or with peptide substrate.
2) Continue to finish calculations for SIRT3/Ac-CS2/NAD+/Methyl-NAM MD simulation and MM-PB(GB)SA calculations.
RC (3/27/14): We will aim to arrange a meeting sometime soon to discuss next steps (including SIRT1 structure prep, discussion of recently posted C pocket structure analysis vis-a-vis project 1, and possibly: residue contribution analysis of MD/MM-GB(PB)SA results and software development plans for automation of virtual screening, binding affinity calculations). In the meantime, please complete those tasks on the list that do not require discussion, including the market survey of rapid cycle PCR (existing software, hardware and users, including features of real-time PCR software such as that on our machine).
Please provide estimated completion dates for these tasks.
PL(03/27/14): 1) C pocket structural analysis is considered done. More input is needed to direct further analysis.
2) SIRT3/Ac-CS2/NAD+/Methyl-NAM MD simulation is finished. MM-PB(GB)SA calculations are currently running, should finish tomorrow.
3) Survey of rapid cycle PCR and related topic is underway, and will be the focus of next week. A report will be generated by the end of next week.
RC: Please list the related topics mentioned that will be covered in the survey, and whether this will occupy all of next week.
I assume residue analysis that was underway at time of paper submission is complete?
PL(03/27/14): I did residue analysis for some of the binding affinities of NAD+ in different systems, including the SIRT3 ternary complex, SIRT3 complexes with NAM or isoNAM, Sir2TM ternary complex and Sir2TM complex with NAM . Only the substrate (NAD+) interactions (vdW and electrostatic) with each residues are calculated.
I will post the list of topics after all the materials get organized, which should include the PCR probe and primer design software, rapid cycler PCR implementation and products by different vendors, recent development of ultra-fast rapid cycler PCR, features of our own PCR machine, and some information about the related PCR market I found from different sources. There are a lot of vendors have products in the rapid cycler PCR or fast PCR market, including enzymes. reagents, assays, thermal cyclers, etc. And some techniques, such as those developed by Fluidigm, I am not sure whether I should add it to the report.
PL(03/17/14): Weekly schedule (03/17/14-03/21/14):
1) Continue to work on assigned tasks:
i.e. structural analysis of C pockets on SIRT3/SIRT1 under different conditions,
h) Next paper (after all the above are done): Begin to set up SIRT3 C pocket MD binding affinity calculation for the tightest binding small molecule identified by XG (for which charge fitting has already been done)
i) Next paper ("): Comment on the comparison of MD trajectories for NAM in Sir2Tm vs SIRT3, with respect to the role of Met70.
j) Next paper ("): Begin to set up NAM/isoNAM C pocket MD simulations for SIRT1. Further details will be discussed when you are ready.
2) Survey on PARP structures and its complexes with PARP inhibitors
PL(03/21/14): Survey of PARP sturctures are underway. Here is an updated version. PARP family.docxWill continue working on the structural survey.
RC (3/21/14): Ok, at some point a comparison of sirtuin/PARP NAD+ binding pockets would be useful. If it takes time, not a priority relative to the other listed tasks on sirtuins. Do PARP enzymes have a Rossmann fold?
PL(03/24/14): Yes. PARP enzymes do have a Rossmann fold. In fact, the Rossmann fold is a protein structural motif found in proteins that bind nucleotides, especially the cofactor NAD.
MD simulation of SIRT3/Ac-CS2/NAD+/Methyl-NAM is under way, should complete by the end of next week. (including MM-PB(GB)SA calculations).
RC: Where on the wiki can we find the list of C pocket ligands that have been tested experimentally with SIRT3 (of which Methyl-NAM is one), along with single structure MM-GBSA correlations?
PL(03/21/14): The correlations with Eric's results can be found under "Task list from Simulations". I also did a correlation plot using 4BVG (SIRT3 with intermediate as receptor) for those small molecules. The plot can be found in the file of Sirtuin_Project01_Week1.docx under "Sirtuin project 1".
Structural comparison of C pocket of SIRT3/SIRT1 started, but not complete yet.
PL(03/21/14): Raj, As for SIRT1 simulations, we have limited structures available.
4KXQ; 4IF6 (SIRT1/ADPR) closed form;
4I5I (SIRT1/NAD+/Ex-527);
4IG9 (SIRT1) open form.
In order to build ternary and quaternary complex, there are more steps and approximations need to carry out. Do you want the NAM/isoNAM in complex with SIRT1 in the binary form or you want to include a peptide substrate and NAD+ as well in the complex? I will try to set them up accordingly.
RC (3/12/14): It would be nice in this context to see some type of structural analysis that shows the effect of peptide binding on C pockets of sirtuin enzymes, which we previously discussed only briefly. This could be provided in the case of SIRT3 or Sir2. This is related to the goal of testing the rank ordering of leads identified so far for SIRT3 C pocket ligands against SIRT1. We would like to have an idea of the order of magnitude of the effects of peptide binding, and hence the aforementioned approximations, on such calculations. I notice that you have referred to this task above, we can decide on which type of complex to use after that analysis is done.
RC: Please provide updates on the above tasks as they become and available along with expected completion dates of the tasks currently underway.
Please provide this update on the task progress Fri am and also provide the planned completion dates for the tasks currently underway today. Such updates should be given on a weekly basis and anticipated completion dates should be given whenever possible, so that feedback and updated priorities can be provided where necessary. If further discussion is advisable before proceeding on a task, please indicate that.
RC: What was the outcome of literature alerts?
PL: The Google scholar alert can only monitor keywords in the title and there is no update since setting up the alert.
RC: Please provide info on the keywords used and whether all of us will be getting updates. If we don't receive any updates after a couple of weeks, please test it with some other keywords.
PL(03/10/14): Weekly schedule (03/10/14-03/14/14):
1) Continue to work on materials required for PCB manuscript;
2) Continue to work on the analysis requested for next paper (some may required further discussion with Raj);
3) Attending Chemical Computing Group's (CCG) workshop on 03/13/14.
4) Begin prepare report on PCR control and latest fast PCR techniques.
PL(02/28/14): Weekly schedule (03/03/14-03/07/14):
Continue to work on materials required for PCB manuscript;
RC (3/3/14): As a priority, it will be useful to have in one table the pdb entries used as starting structures for each MD simulation, how the ligand complex was prepared (e.g., by alignment with another
structure such as Sir2) and (as a third column) what structures were compared to in each case for rmsd analysis).
Note on the comment made on manuscript revision page regarding highlighting some ligand-residue contacts: some of those contacts that differ between the crystallographic and MD average could be added to the bullet points list that was previously prepared (since from looking at the figs I assume there are many contacts that are preserved between crystallography and MD average).
Once I am able to access the latest figure doc files on dropbox I will review and request edits for the final figs.
PL(02/21/14): Weekly schedule (02/24/14-02/28/14):
1) Prepare materials required for PCB manuscript;
2) Continue MD on Sir2TM/NAD+ binary system and run MM-PB(GB)SA calculation;
3) Run MD on Sir2TM systems and Ac-p53 systems to produce MM-PB(GB)SA energies for free protein and ligand
4) Continue survey on Rapid Cycle PCR and fast PCR and begin report preparation
5) Continue small molecule docking for SIRT3 in C pockets using Glide and MM-GBSA (Prime)
RC (2/23/14): Comments regarding schedule/tasks (breakdown of tasks under 1 above, which is priority):
a) Figures: NAM/isoNAM ligand interaction diagrams needed.
Edits to other figures may be requested shortly.
b) Manuscript discussion: discussion will consider relative binding affinities of NAD+ and NAM, in SIRT3 and Sir2 AC pockets of ternary complexes. Provide bullet points comparing these binding affinities and the relevant interactions visible in the average MD figures as they relate to the MM-PB(GB)SA energy breakdown by interaction type. This will be used to replace the discussion written by Eric on the comparison of protein:NAD+interactions in SIRT3 and Sir2.
c) Manuscript discussion: related to b), for each of the complexes for which binding affinities are reported and which were discussed in the original paper draft, provide bullet points regarding the relevant interactions visible in the figures as they relate to the MM-PB(GB)SA energy breakdown by interaction type.
d) Manuscript discussion: Provide bullet points comparing the fluctuations in the MD trajectories for NAD+ in the SIRT3 and Sir2Tm ternary complexes (relt'd to b). If entropy was calculated for the complexes which equilibrated, this could be used as a point of comparison of the distributions.
e) Manuscript discussion: Provide the requested average MD figure for a snapshot other than the last 10 ps which shows another mode of the distribution, in order to assess how much discussion should be devoted to the interactions in the average MD figures.
f) Methods: Pending methods sections should be completed
g) Methods: Please confirm that entropy is included in the reported binding affinities. MD convergence analysis: would like more details to be provided on how simulation time was set/how convergence of MM-PB(GB)SA calculations was assessed (or a figure with caption to be prepared showing the convergence rate of the MM-PB(GB)SA binding affinities with simulation time for a couple of complexes)
h) Next paper (after all the above are done): Begin to set up SIRT3 C pocket MD binding affinity calculation for the tightest binding small molecule identified by XG (for which charge fitting has already been done)
i) Next paper ("): Comment on the comparison of MD trajectories for NAM in Sir2Tm vs SIRT3, with respect to the role of Met70.
j) Next paper ("): Begin to set up NAM/isoNAM C pocket MD simulations for SIRT1. Further details will be discussed when you are ready.
PL(02/14/14): Weekly schedule (02/17/14-02/21/14):
1) Run MM-PB(GB)SA calculations on Sir2TM/peptide/NAD+ and Sir2TM/peptide/NAD+/NAM systems;
2) Prepare requested figures for PCB manuscripts;
3) continue to finish the computational method section.
PL(02/07/14): weekly schedule (02/10/14-02/14/14):
1) Continue MD simulation for Sir2TM built upon 2H4F, including both Sir2TM/peptide/NAD+ and Sir2TM/peptide/NAD+/NAM system;
2) Component analysis for MM-PB(GB)SA results;
3) Continue survey for Rapid Cycle PCR, high throughput PCR and PCR software and their applications
4) Continue the draft for MD/MM-PB(GB)SA methods
Raj, can you highlight the questions that you think are important for the PCB manuscript and you are still waiting for answers?
RC (2/9/14): Figures needed for the PCB manuscript:
- 3D representations. Superimpose crystal structure and report RMSD of ligand (MD average to crystallographic) wherever possible. Complete captions indicating method of preparation
n SIRT3:peptide:NAD AX and AC pocket MD averaged structures (parts a,b of figure) and SIRT3:NAD AC pocket (part c of figure)
n SIRT3:NAM/isoNAM: superimposed averaged MD structures in part a of figure. Sir2Tm:NAM averaged MD structure compared to crystallographic structure in part b of figure. Compare binding affinities for both NAM and isoNAM in SIRT3, indicating NAM,isoNAM are congeneric series and binding affinities can be compared
n Sir2:peptide:NAD AC pocket averaged MD structure, comparison to crystal structure.
- Ligand interaction diagrams for all the above (MD average only). Please confirm that all the ligand interaction diagrams prepared by Eric have a corresponding figure based on MD simulations. Binding affinities and energy component analysis will be requested thereafter.
RC (2/14): a) Draft of the MD/MM-PB(GB)SA methods should be submitted today.
b) Draft figures from the list above that are complete should be submitted today. If you have any questions regarding the preparation of these figures please let me know. As noted these are the priority and needed details of the other analyses will be provided after the above are received.
PL(02/04/14): weekly schedule (02/03/14-02/07/14):
1) Analyze MD results for manuscripts
RC: Please provide more details on this so I can get an idea of the time required.
b) Reply to all remaining questions on PCB Manuscript Revision page.
PL (02/04/14): It is common practice that after MD simulations, we look into the trajectories and figure out what is the change in the MD and if there is any significant points that we can derive. And sometimes, structural fluctuations, hydrogen-bonding network in ligand binding site, need to be analyzed. And we also did MM-PB(GB)SA calculations, and we might want to related the energy terms to the structural contacts from trajectories. Usually, MD simulations are able to reveal more information that is not available from crystal structure as the later only provide a still picture at low temperature. And the time required varies.
RC (2/4/14): I am aware of common practices. I am referring to whether you have finished all MM-PB(GB)SA calculations for all MD simulations run to date and whether you are referring to MM-PB(GB)SA calculations as analysis of MD.
2) Schedule new MD simulation if needed
3) Survey for rapid cycle PCR, fast PCR and high throughput PCR and potential use of PCR optimal control software.
RC: This is approved.
4) Writing program for MM-GBSA calculation using Ambertools from docked pose obtained by Glide (to avoid the license fee for Prime in the future.)
RC: We will have to review/agree on the plan for this before proceeding.
RC (2/3/14): PL should use this page to post his weekly schedule at the start of each week (including this week).
.
PL (01/21/14):
Focus on week 01/20/14-01/24/14:
1) Carry out survey on PCR software, check out PCR software control we currently use.
2) Search for patents on PCR software.
3) Survey for customers for high-throughput and fast PCR.
4) Summarize on Primer design and their commercial customers.
RC (1/29): Please post here the original email sent to PL regarding the plan for PCR software. Background material has been posted to the COLD PCR page.
Please pay special attention to 3) the market for rapid cycle PCR amplification, since it is related to our technology for minimal time PCR. There are many hardware designs that claim to be capable
of rapid cycle PCR. We may want to contact some of these companies to determine if they are interested in showing that their hardware can support minimal time PCR protocols computed through our algorithms. So under 3) we should identify both the customers of high-throughput/fast PCR as well as the companies that are making the devices that these customers are buying.
Email to PL (01/15/2014):
Ping,
As discussed, one part of your job description will be scientific software development. Before developing any software packages, it is important to assess the competition and customer demand.
The first assignment in this regard is to evaluate existing PCR software technology and analyze the market for PCR software products.
I would like you to work on this whenever you do not have any immediate pending tasks for the sirtuin project (e.g., if you are waiting for a simulation to run).
-- Our technologies:
a) primer rate constant prediction,
b) minimum extension time calculations,
c) temperature cycling protocol prediction - both single sequence and multiple sequence discrimination (e.g., COLD PCR)
Tasks:
1) Produce a comprehensive list of all major commercial primer design and pcr software, with a summary of the capabilities of each.
2) Classification of the above by a) company and b) hardware requirements and software dependencies: does the software require a pcr machine manufactured by the same company? Is it part of a larger software platform?
3) Who is the software developer? i.e., is the software developed in-house or by a third party? (if info is available; may require communication with company reps)
4) Who is the end user? research labs, clinical diagnostic laboratories, sequencing facilities,...?
5) Who can be contacted for more information?
6) List of patents, if any, pertaining to primer design or pcr cycling protocol prediction
Examples: a) fluidigm is advertising a microfluidic pcr machine that can cut cycling time 3x for high-throughput genotyping applications.
They may be interested in integrating our algorithms that minimize cycle time with their existing software platforms for further improvements.
Who might be the primary customers for this technology?
b) current real-time pcr software shipped with pcr machines (e.g., CFX from biorad) provide temperature cycling protocol recommendations but these are not optimal and do not provide automated prescriptions for diagnostic applications (e.g., COLD PCR protocols for mutation detection).
Also, their primer design software only calculates Tm's, not reaction rates, and hence cannot be used for reaction time optimization.
c) Life Technologies/Applied Biosystems has QuantStudio and Primer Express software for real-time PCR and primer design, respectively.
In the process of evaluating these issues it may be necessary to contact/communicate with companies that sell pcr-related software, either by email or phone.
As you know PMC-AT is not a single biotech startup. Rather, it is a technology investment firm that incubates and invests in the development of technology in several areas of biotech. In this capacity, we are often approached by biotech diagnostics or therapeutics startups looking for funding. Since these companies generally do substantial market analysis in their product areas, we can learn from them about the market demand for our products under development. Also, they may constitute potential partners who can assist with product marketing and distribution. I may ask you to attend either a phone call or presentation by these companies from time to time.
Based on this analysis we will decide whether your first software development project will be for pcr or another application.
We can meet to discuss this at a convenient time, once you have had a chance to digest the above.
Raj
CJ (12/30/2013)
I checked the JMB 1971 paper by M.E. Craig about the kinetic experiments: they used a temperature jump apparatus that is customized with a UV detector. After a temperature jump, the UV absorption is monitored at micro-second to ms time scale. The change in UV absorbance is only ~0.01, therefore the UV spectrometer they used must be extremely sensitive and accurate. Apparently this kind of experiment can not be performed with any instrument currently in our lab. I roughly searched on internet for temperature jump apparatus with UV detector, but did not find anything commercially available. I guess because this kind of instrument is not widely used, we need to buy a temperature jumper and a UV detector separately, and then try to conjugate them together. Please let me know if we are still interested in it. If so, I can look for more details.
RC (12/17):
PL, please work on the following PCR tasks until I provide further feedback:
-Determine whether MELTSIM is open source and what language it is written in, in case we want work with the code in new applications; in that case we would translate and integrate current matlab code with it.
-Get list of mutations for COLD-PCR from CJ (including mutations that were looked at by Sudha, if available) and apply MELTSIM to compute the melting curves. First list the mutations to be studied. Compare the results to Poland software that CJ worked with.
-Read the JCP paper, especially the part that uses experimental relaxation time data to estimate the parameters in the model. This requires experiments (e.g temperature jump experiments) to measure the relaxation times. Ask CJ about the protocol/equipment required for this. The computational part of the work would be to apply least squares estimation with the experimental data and the model.
-Comment on whether the codes in the JCP codes folder just shared via dropbox are sufficiently clear vis-a-vis the results they were used to generate in the JCP paper (e.g. on relaxation time for heterogeneous sequences).
Let's post general information regarding our papers to pages other than the PL tasks page.
PL(12/18): MELTSIM is open source, and the only version available right now is for unix/linux, it is written in C. I will work with CJ on COLD-PCR data and simulation.
I will look into the codes you shared, read JCP paper and work with CJ on the data modeling and fitting.
XG(12/17): Dr. Sinclair with sirtris group had published one paper titled as " Biochemical characterization, localization, and tissue distribution of the longer form of mouse SIRT3". In the paper, it was reported that NAM inhibits mouse SIRT3 competitively respect to NAD.
The abstract is listed below:
SIRT3 is a key mitochondrial protein deacetylase proposed to play key roles in regulating mitochondrial metabolism but there has been considerable debate about its actual size, the sequences required for activity, and its subcellular localization. A previously cloned mouse SIRT3 has high sequence similarity with the C-terminus of human SIRT3 but lacks an N-terminal mitochondrial targeting sequence and has no detectable deacetylation activity in vitro. Using 5′ rapid amplification of cDNA ends, we cloned the entire sequence of mouse SIRT3, as well as rat and rabbit SIRT3. Importantly, we find that full-length SIRT3 protein localizes exclusively to the mitochondria, in contrast to reports of SIRT3 localization to the nucleus. We demonstrate that SIRT3 has no deacetylation activity in vitro unless the protein is truncated, consistent with human SIRT3. In addition, we determined the inhibition constants and mechanism of action for nicotinamide and a small molecule SIRT3 inhibitor against active mouse SIRT3 and show that the mechanisms are different for the two compounds with respect to peptide substrate and NAD+. Thus, identification and characterization of the actual SIRT3 sequence should help resolve the debate about the nature of mouse SIRT3 and identify new mechanisms to modulate enzymatic activity.
The inhibition mechanism was not discussed in the paper.
PL (12/17): During the course of the manuscript reviewing and modification, as well as calculations carried out to complement the manuscript, I found that the biggest issues is actually not reviewer's critics, it is the overlooking of some prior experimental results from other groups that is critical to our interpretation of experimental finding and computational design. Here is a critical review on Sitruin chemical mechanisms that covers many important aspect of the what we discussed in the manuscript. 1-s2.0-S1570963910000361-main.pdf
This review contains many valuable information that can help us understand sirtuins and their chemistry better, and is worthy of complete digestion.
Here is the list of the some key information from the reviews and the references there:
1) Section 2: NAD+ binding gemetry and affinities: Km of NAD+ for Sir2 (29-70 uM), SIRT1 (132-550 uM). For comparison, Km of NAD+ for SIRT3 was reported to be ~600 uM. For comparison, AceCS2 has a binding affinity of 15 uM. [ Jin, L. et al. Crystal structures of human SIRT3 displaying substrate-induced conformational changes. J. Biol. Chem. 284, 24394–405 (2009)], our single structure MM-GBSA scores failed to differentiate the Km of NAD+ for Sir2 and SIRT3.
2) Section 5: Partitioning of the intermediate (nature of nicotinamide inhibition): Strong correlation between base-exchange and nicotinamide was well established, there are also many discussions of various findings. Our prior kinetic model relied simply on the traditional competitive/non-competitive inhibition and failed to address the inhibition mode through base-exchange as confirmed by experiments. And our design of computational study provided is based on a incomplete or maybe false kinetic model.
3) Section 6: Nicotinamide derepression: Brief discussion of iso-nicotinamide effect and references are provided. And apparently, iso-nicotinamide's derepression effect is achieved through blocking the base-exchange reaction by nicotinamide. We have experimental results showing that iso-nicotinamide by itself, (without the presence of nicotinamide), is a weak inhibitor with IC50 of 13.8 mM. (data not in the manuscript.)
One interesting finding of our experimental result is that SIRT3 displays an apparent competitive inhibition behavior, which differs from findings in many other Sir or sirtuins, and is also against the claim by A. A. Sauve's (author above) claim that inhibition through base-exchange is expected to produce non-competitive inhibition.
Xiangying and I have tried to complete the fit the data to the more completed kinetic model and hopefully we can have a correct interpretation of our data and finding, and find out what is needed to support our finding. We found that our data can fit decently using base-exchange only inhibition mechanism, and the apparent competitive behavior can be achieved when NAD+ has a weak binding affinity. The current MD and MM-PBSA result seems to support such finding, and the ITC results by Jin et al (see reference above) also support such finding. With a more completed model, which takes into account of both NAM binding in the C pocket and base-exchange mechanism, we can achieve better fitting to the data. And we find it makes a decent story and we can use the the more robust kinetics model to help answer some questions raised by the author in Section 5 above.
Compare to the questions raised by the reviewers, I found these questions is more critical to the quality of our work.
I would suggest we have a meeting to discuss what we should do for the manuscript.
RC (12/13):
--Thanks for providing some updates regarding replies to some of the reviewer comments on the project 1 page. Based on that document it seems that several of the questions are still unanswered and that we may not have started making changes to the manuscript yet per the findings below. Please see the comments below (esp 11/28) regarding the detailed priorities for the paper resubmission vis-à-vis required computational revisions. Please send me the latest paper draft with the edits indicated below. That version of the draft will later be augmented with the response to reviewer 1’s MD comments.
-- After the B pocket MD simulation proposed below, we would consider the readiness of the paper for resubmission. At that time we can for example discuss whether further MD simulations are required. Please see some additional comments made to the paper 1 page regarding MD.
-- After finishing the above paper edits (if not already complete) we will start some work on PCR project next steps, which I will send shortly, alongside continued work on virtual screening for paper 2 and MD simulations.
-- On a related matter: regarding Schrodinger licenses, the non-academic pricing we recently received for the current 24 token license is very expensive. I am asking Carrie for pricing on a Glide-only license. In that case, we may have to use non-Schrodinger tools for everything other than docking. Please let me know if you see any issue with this.
PL(12/13): As I presented on 11/22, in order to answer reviewer 2's questions, I redo the "in place" MM-GBSA calculation for NAD+ in 1YC2 (all four chains used, NAM were kept in place for AB pocket binding), and in 4FVT.
NAD+ AC binding in Sir2 (1YC2 B/C chain): -89.8 and -92.5
NAD+ AB binding in Sir2 (1YC2 A/D chain): -58.8 and -85.9
NAD+ AC binding in SIRT3 (4FVT with AceCS2): -99.87
NAD+ AX binding in SIRT3 (4FVT with AceCS2, MM-GBSA score after docking with Glide SP): -81.4 (X pocket is not exactly the B pocket, but close.)
These results actually do not support our conclusion and our explanation for competitive inhibition mode in SIRT3.
RC (12-13): Please see below. As noted these results will not be interpreted in terms of an inhibition mode for SIRT3. That will be done via MD simulation. As specified below on 11/28 several of the other comments could be addressed using these results and other answers we have prepared together, and the priority should be answering those questions (both experimental and computational) asked by the reviewers directly in the manuscript. If not please indicate directly below my comments from a couple of weeks ago why not, as I did not receive a reply to these comments and was under the impression that changes were being made per our discussions.
PL(12/13): Minor modifications, such as details on the in-place MM-GBSA calculations, the selections of chains were made into the manuscript. However, concerns brought up by reviewer, were not answered, such as for section "Computational Modeling of NAD+-SIRT3 binding" 2)" It is not clear from the Methods whether the peptide substrate is cleaved (or removed) from the binding site before NAD+ docking. If it is not removed, would the docking results still make sense? Can the induced-fit docking method handle two disconnected proteins?"
As you suggested on 11/28, I did rescore NAD+ binding in AB pockets (the result presented above), did find out the detailed settings used by Eric, and made changes to the manuscript to answer the questions by reviewer 2. However, the changes are not complete as we still need to combine MD simulation (requested by reviewer 1) results in the study.
I will combine my changes with Xiangying's updated manuscript when she return next Monday.
RC(12/16): As noted below on 11/28, at least two of the major concerns of the reviewer - robustness to choice of crystal structure and similarity of AB/AC pocket binding scores for Sir2 vis-a-vis noncompetitive inhibition - can be explained by the rescored results. Also as noted it appears that we can provide more methodological details now (now that we have understood the reason for the lack of robustness of Eric's results). This is the first time I have seen the AX pocket binding rescore, and so I am not sure about the methods used to obtain that structure (below on 11/28 I was referring to the score for the template induced fit structure, which is not a substitute for MD; I am not sure that is what you are reporting above). The other points above are not as central to manuscript's conclusions.
PL(12/18): I believed the reviewer's comment on confidence in computational results is intent for more details on the setting and choice of methods. And I think the extra details I put forward should be able to answer the reviewer's question. And yes, the binding scores we had can be used to explain the choice of crystal structure. And as many believe that base-exchange will lead to a noncompetitive like inhibition, a similar scores for AB/AC pocket binding probably won't change the behavior.
As to the AX pocket binding, I first showed the result during the group meeting presentation on 10/19. The structure is obtained by first docking NAM into the C pocket of 4FVT structure, followed by docking NAD+ into receptor of SIRT3/AceCs2/NAM complex. Eric's attempt previously failed may be due to the minimization at protein preparation, not removing extra heteroatoms, C pocket not pre-occupied, and ranking of NAD+ in AB binding mode fall to low to be included in the output. Eric has various MM-GBSA scores for pose from template-induced fit. I am not sure what you mean by rescore NAD+ in the SIRT3 AB pocket? Do you mean we rescore it after MD simulation?
RC (12/16): I will look closely over the revised manuscript after receiving confirmation that all the points listed above have been directly addressed in the paper. Please confirm. Thanks.
One of my concern with our current computational study and conclusions is that we designed out study based on traditional competitive, non-competitive inhibition mode and didn't take into the consideration of the inhibition effect from base-exchange reactions. And from the values I am getting, I am not sure we have the correct answer to the inhibition mechanism.
RC (12/16): I have prepared some commentary on different inhibition modes and their role vis-a-vis this paper, and I will provide them after receiving and reviewing manuscript changes regarding all other reviewer comments we have discussed to date except MD. One point to note for now: To determine the extent to which the observed inhibition kinetics mode for SIRT3 inhibition by NAM (inhibitor=product) is affected by base exchange inhibition, one could do the MM experiments for another inhibitor (where inhibitor is not equal to product and hence does not engage in base exchange), such as isoNAM. These isoNAM experiments were therefore requested over the last 6-9 months but have not yet been done. A suggestion was made at one the latest meetings that this and alternate kinetic models should be considered in a subsequent paper, and hence the experiments were not prioritized over the last month.
As I understand it the points for discussion regarding the inhibition modes are briefly as follows:
a) The latest data regarding the binding affinity of NAD+ to a pocket other than the AC pocket (X pocket) and its implications for a standard picture of competitive inhibition.
b) A generalized model including base exchange inhibition as well as the possibility of binding to an alternate pocket (X pocket) can fit the data (for NAM) equally well or perhaps somewhat better than the standard competitive model
c) The appropriate way to present the inhibition mode section of the paper for SIRT3 to accommodate the possibility of binding to the alternate pocket
Please confirm, and if there are any other major points for discussion, please list them briefly.
I have carried out a 16 ns MD simulation for AB pocket binding mode. The system was constructed by supper-impose NAD+ and NAM on to 4FVT structure, and we found that NAD+ in AB binding mode (not exactly the same but close) remains stable after relatively long simulation.
A limited MM-PBSA calculation (without entropy correction) from MD simulations do suggest that NAD+ binding with SIRT3 is much weaker compare to acetylated peptide (AceCS2). And here is a rough result (from 300 frames) Delta H (binding, AceCS2): -10.93 +/- 3.9 kcal/mol, Delta H (binding, NAD+): -3.03 +/- 6.45 kcal/mol, Delta H (binding, NAM): -0.34 +/- 3.0 kcal/mol,
Analysis of Xiangying's data using the new kinetic model based on base-exchange reaction suggested that the competitive-like Lineweaver–Burk plot (or double reciprocal plot) may be a result of weak NAD+ binding in SIRT3. I suggested we investigate further on this project.
1) The posted update on manuscript by Xiangying was responses to reviewer's comments. The modified manuscript was not uploaded.
2) I can start work on PCR project while continuing virtual screening.
3) Please also ask Carrie for pricing on both Glide and Prime licenses, Protein Preparation and Ligand preparation wizards are also needed if the license is not covered by Glide or Prime.
RC(12-17): Updates from Carrie on licenses:
"One license each of Prime, Glide, LigPrep, and Epik is $28,000. This includes Induced Fit Docking (Glide + Prime), Protein Prep Wizard (Epik),
and ligand preparation (LigPrep)."
"Here are some additional options that include LigPrep and Epik:
- one license each of Glide, LigPrep, and Epik is $16,000
- 10 tokens with Glide, LigPrep, and Epik is $24,000
- 16 tokens with Prime, Glide, LigPrep, and Epik is $38,400 (having Prime and Glide lets you run the Induced Fit Docking Workflow)"
"I just spoke with my boss again, and in an effort to help you transition to the new licensing set up, I can offer you a 50% discount on the 25-token
library for the Small Molecule Drug Discovery Suite ($30k total)."
"One license of Glide alone is $12,000 for one year."
We will need to assess the need for token vs single license packages.
RC (11/21): I'm posting here regarding computational follow up tasks for the 1st sirtuin paper review, as well as hardware/software issues. Please feel free to move these discussions to other pages if you
deem fit.
Comments on MD simulations for 1st sirtuin paper revisions:
- Certainly MD can help overcome issues with insufficient conformational sampling. We chose to submit first version without extensive MD, with plan of revisiting these issues upon review.
- Long MD simulations are more appropriate for mechanistic studies in this paper than for high-throughput lead discovery drug screening
- However must be aware of errors in MD binding affinity estimates. So it is important not to expect too much from it in terms of binding affinity estimates or to assume from the outset that all structures reported in this paper should be reassessed w MD. We do not have direct access to experimental binding affinities of NAD+ and are not looking at a congeneric series; hence we would have rely on known correlations between MD/MM-GBSA and LIE to support our contentions regarding the relative magnitudes of binding affinity estimates for different binding modes. Apparently, MD/MM-GBSA and LIE display similar correlation to experimental datasets (please verify, since LIE uses more regression factors).
- Important: Reviewers will likely not be ok treating a paper where nearly all computational sections are rewritten as the same paper
- Hence need to prioritize those simulations that are most relevant to the paper's message. For example, using template-based superposition to locate a starting structure for NAD+ in the SIRT3 AB pocket, followed by equilibration and determination whether NAD+ remains in that pocket. We should start with these high priority simulations, and depending on results, decide how far to continue with other MD simulations before resubmitting the paper. PL to provide rank ordering of simulations by priority. Other MD simulations could be continued even after paper resubmission depending on available time given work on other projects.
(For example, the relative binding affinities reported for AB/AC pockets of SIRT3 are the most suspect because they do not include conformational rearrangement energy. If we can show more rigorously with MD that SIRT3 AB pocket binding is highly unfavorable, we might choose keep the Sir2 AB/AC pocket single structure binding affinities in the paper. since we could then justify the statement that NAD+ binding to both pockets of Sir2 is favorable, whereas this is not the case for SIRT3. This is one of the primary arguments of the paper.)
- RC would like to see some validation of MD equilibration resulting in stationary distribution that is largely independent of starting conditions/structure, by comparing across several starting crystal structures (e.g. monomers from the Sir2 multimer crystal structure). This was a concern of reviewers. Propose a benchmark using selected structures. Ideally this could be done with a shorter duration MD simulation, not a full binding affinity calculation on each structure. PL to provide more info on proposed benchmark.
- Based on this week's update on referee report answers, RC did not have a good idea of whether the salient conclusions of the MM-GBSA scoring were called into question by the lack of robustness to choice of crystal structure. Additional details on which conclusions are suspect is needed. PL provided more info verbally, please repeat comments here.
- PL mentioned some of EK's docked structures are not appropriate starting structures for MD binding affinity calculations due to other molecules being present. PL to start by recomputing single structure binding affinities after removing the extraneous molecules used for crystallization. If the estimates are more robust to variation in the crystal structures that EK's were, the MM-GBSA estimates may be sufficient for this paper. If not, we should proceed with the MD equilibration benchmark above as the next step.
RC (11/28): Based on PL's most recent results below, single structure MM-GBSA estimates for Sir2 AB/AC pockets may indeed be sufficient for this paper. We should be able to answer many of reviewer 2's computational questions now - in particular, the ones about what we consider to be roughly similar scores in a noncompetitive inhibition model, and the robustness of MM-GBSA scores to change of starting structure. Let's answer these questions and identify what questions from reviewer 2 remain unanswered. I believe there are some remaining questions regarding more detail on computational methods; it appears it is worthwhile providing these now? Should we also rescore NAD+ in the SIRT3 AB pocket, to check the relative binding affinity with respect to the SIRT3 AC pocket score below? After that, it appears that the priority for MD simulation is then NAD+ in the AB pocket of SIRT3.
PL(11/22): New In-place MM-GBSA calculations using 1YC2 (chain A B C D) and 4FVT were obtained under such setting: All not relevant molecules (i.e. PEG, EOL, extra Zn^2+, SO4^2-, and waters were removed, hydogen assigned based on pKa estimation, only hydrogens were optimized, NAM were kept in 1YC2 chain A and D, Acetylated peptide was kept in 4FVT) . And here are the MM-GBSA values:
1YC2 (Chain A) (AB pocket, with NAM in C): -85.91
1YC2 (Chain D) (AB Pocket, with NAM in C): -58.75 (Note that the amide group has different orientation compared to chain A. however, this does bring the issue about which structure to use for comparison.)
1YC2 (Chain B) (AC pocket): -89.78
1YC2 (Chain C) (AC pocket): -92.48 (chain B and chain C is very similar in structure, therefore close values are expected)
4FVT (AC pocket, with peptide): -99.87
By removing also NAD in chain A and D. I obtained the MM-GBSA values for NAM in C in 1YC2:
1YC2 (Chain A) (NAM in C): -31.48
1YC2 (Chain D) (NAM in C): -31.12 (again, NAM in chain A and D is very similar)
In term of relative binding affinity, it may be OK to use single structure for comparison.
RC (11/23): Are we looking at MM-GBSA scores for NAM binding to the respective pockets above? I.e., none of the above pertain to NAD+?
E.g., when we say AB pocket, with NAM in C, what does "AB pocket" refer to? I assume this does not mean NAD+ in AB pocket, since NAD+ scores are discussed below.
PL(11/26): The top five scores refer to the NAD+ binding in Sir2 and SIRT3, and the "AB pocket" and "AC pocket" mean NAD+ in those pockets, at the same time, for NAD+ in the AB pocket, NAM always occupies C pockets and, the MM-GBSA values were obtained while keeping NAM in the C pockets.
Without NAM in C pocket, similar values are obtained.
1YC2 (Chain A) (AB pocket): -85.45
1YC2 (Chain D) (AB pocket): -58.30
RC (11/28): Ok, in that case it looks like we have resolved the issue regarding "similar" scores for Sir2 AB/AC pocket binding, raised by the reviewer as well as by us.
However, when I performed docking using Glide XP, (consider it an optimization using Glide scoring function) the MM-GBSA values changed.
For NAD+ binding in C pocket, (1YC2 chain B and C) the best MM-GBSA scores are -103.75 and -99.65; For NAD+ binding in B pocket, the native structure can not be obtained and the best MM-GBSA score obtained are --64.96 and -84.78 for 1YC2 chain A and D.
RC (11/28): I understand the above as being Glide XP values.
- Regarding the base exchange mechanism not being studied in this paper: RC mentioned that Wolberger did not appear to highlight base exchange in her description of noncompetitive inhibition in her structure papers.PL indicated that the role of base exchange was mentioned by Wolberger. PL can indicate where this was discussed in the Wolberger paper.
PL(11/22) In Wolberger's Molecular Cell paper (attached, with 1YC2 structure) 1-s2.0-S1097276505011214-main.pdf, on page 1, last paragraph, the first sentence states, " Nicotinamide inhibits the deacetylation activity of sir- tuins by reacting with a reaction intermediate." She went on to provide the reference paper Sauve published in 2003 and Jackson et al published in 2003, both provided strong evidence that nicotinamide inhibition occur via base-exchange reaction.
RC (11/24): Since both Sir2 and SIRT3 undergo base exchange inhibition, what then would be the explanation of the observed differences in inhibition kinetics if indeed base exchange is the primary source of inhibition? Why would the inhibition kinetics of the two enzymes fit the standard noncompetitive/competitive inhibition models in that case? XG, please remind me of the literature we looked at when considering Sir2 inhibition noncompetitive due to the AC pocket binding. If there was literature that showed noncompetitive inhibition of Sir2 via MM kinetic fitting, did it explicitly refer to AC pocket binding, or perhaps just imply it?
XG(11-25): Sinclair’s group (JBC _2002_277: 45099-45107.) has shown that nicotinamide strongly inhibits the deacetylase activity of both yeast Sir2 and the human homologue, SIRT1 in vitro by MM kinetic fitting.
Data are shown as a Lineweaver-Burk double-reciprocal plot of 1/v vs. 1/[NAD+].
They also propose a model for Sir2 regulation by nicotinamide where noncompetitive binding refer to AC, AB pocket binding.
Sir2-catalyzed deacetylation consists of two hydrolysis steps that are thought to be coupled. Cleavage of the glycosidic bond connecting nicotinamide to the ADP-ribose moiety of NAD+is followed by cleavage of the C-N bond between an acetyl group and lysine. Min et al from Cold Spring Harbor Laboratory reported the NAD+binding pocket of Sir2 enzymes contains three spatially distinct sites (A, B, and C), the later two of which are thought to be directly involved in catalysis (A _Crystal Structure of a Sir2 Homology-NAD Complex (2001) Cell 105: 269-279). In the presence of an acetyllysine, NAD+bound to the B site can undergo a conformational change bringing the nicotinamide group in proximity to the C site, where it may be cleaved (B). The ADP-ribose product of this reaction may then return to the B site where deacetylation of the acetyllysine occurs. They propose that nicotinamide binds to and blocks the internal C site, preventing the conformational change and subsequent cleavage of NAD+(C).
References:
Crystal structure of a SIR2 Homolog-NAD complex_2001_Min.pdf Inhibition of Silencing and accelerated aging by nicotinamide_2002_Bitterman.pdf
We will be doing NAM/iso-NAM MD with intermediate for paper 2 anyway, so we should retain the original schedule for the NAM/iso-NAM binding affinity calculations in the presence of the intermediate. After making more progress with virtual screening, PL will have a better idea of when MD for paper 2 will start.
PL (11-20):
Raj,
First, I want to emphasize that current project discuss some interesting aspects on modulation of sirtuin activity by small molecule, NAM and
iso-NAM, which reviewers agreed that, with improvement on the computational study, it can be published on PCB.
However, I am afraid that simply providing more details of the docking can only expose the problem in the current study, and I myself do not
have confidence that current results can provide convincing support to our conclusion.
From my own opinion, we need to redesign the whole computational study instead of following up with what Eric has had. And I list the reasons
below:
1) The docked complex structures from Eric is not a standard way in preparing the starting structure for MD simulation, the extra
co-crystalized molecules are not very relevant and should be removed. And I can easily prepare the starting structures for the simulation that
will be more consistent.
2) With new SIRT1 crystal structure, we can use these structures to prepare for the MD simulation, which directly relate to the experimental
results we are having.
3) Our current study failed to address the base-exchange reaction and its role in inhibition by NAM. The current study's interpretation of non-competitive and competitive
behavior actually contradict what other people have found that base-exchange is probably the major cause for inhibition by NAM, at least for Sir2. Docking NAM and iso-NAM
into SIRT1/SIRT3 with intermediate followed up with MD simulation will help build a more complete picture of the mechanism. 4) Computational studies, including MD results
may demand different explanation and draw different conclusions we currently have. Therefore it is not necessary we stick to Eric the results right now.
For the reasons above, I suggest not the put too much effort into revising the manuscript right now and wait for the outcome of the MD
simulation before proceeding further. And I also suggest to start from scratch to build the models for simulation instead of following up on
Eric's results. I mentioned in the last meeting that the minimum MD simulation required
as followed:
Reactive ternary complex: Sir2(or SIRT1)/acetylated peptide/NAD+(in AC
pocket)
Non-reactive ternary complex: Sir2(or SIRT1)/acetylated peptide/NAD+(in
AB pocket)/NAM(in C)
Reactive ternary complex: SIRT3/acetylated peptide/NAD+(in AC pocket)
Non-reactive ternary complex: SIRT3/acetylated peptide/NAD+(in AB
pocket) /NAM(in C)
Binding free energies for NAD+ will be evaluated and compared.
And I would also suggest we run extra simulations:
SIRT3/Intermediate /iso-NAM(in C)
SIRT3/Intermediate / NAM(in C)
Binding free energies for iso-NAM and NAM will be evaluated and compared.
MD benchmark simulations are rather standard.
Amber 99SB force field is used.
For Zn, extra parameters are from the following reference:
1) Lu, Q., Tan, Y., Luo, R., Molecular Dynamics of p53 DNA Binding
Domain, J. Phys. Chem. 2007, 111:11538-11545
NAD+ was obtained from the following reference.
2) Walker, R.C, de Souza, M.M., Mercer, I.P., Gould, I.R., Klug, D.R.,
J. Phys. Chem. B., 2002, 106(44), 11658-11665
3) Pavelites, J.J., Gao, J.L., Bash, P.A., Mackerell, A.D., J. Comput.
Chem. 1997, 18, 221
Acetylated lysine was obtained from the following reference.
4) G.V. Papamokos,G. Tziatzos, D.G. Papageorgiou, S.D. Georgatos, A.S.
Politou, E. Kaxiras, Biophysical Journal 102 (2012) 1926-1933.
The initial structure of SIRT3/NAD+/peptide complex was constructed from 4FVT. Using AmberTools, extra 14 Sodium ions and 5 Chloride ions
were added to neutralize the system, then the system was soaked in a box of TIP3P water molecules with a margin of 12 Angstrom. Prior to free MD
simulations, a 10000-step minimization is performed with a constraint of 5 kcal/mol/A^2 on heavy atoms of complexes, followed by a 200 ps NPT MD
under the same constraint. 2 fs time step was applied together with SHAKE algorithm on all bonds involving hydrogens. Langevin dynamics was
used for temperature and pressure control (at 300 K). Electrostatic interactions were computed using Particle Mesh Ewald (PME) method.
Simulations were carried out using NAMD on slave006 node (16 cores). Usually 2ns simulation can be obtained using 16 cores in 24 hours.
I do not have a quick fix to the current computational study. And I believe that it is very worth the effort in order to produce a high
quality paper.
Also, we need to plan ahead for the purchase of new computing facility that meets our needs.
Please let me know your plan on the project.
Thanks,
Ping
RC (11-18):
A high-level summary of some action items from the meeting last week:
1) XG: Start addressing experimental questions from referee reports.
Estimated time: ~ 2 weeks.
2) PL: a) Carefully read the reviews. Determine which computational
questions can be answered now. Revisions that do not require new
computations can be posted to the wiki, along with suggested replies to
referee questions.
b) Review the associated structures generated by Eric for each Figure
(along with associated methods used to generate them), refreshing your
memory on the computations pertaining to each of the referee's
questions. Also, based on this revisiting of the structures in the
paper, consider which ones are of the highest priority for MD
simulations (I will do the latter as well).
Whichever binding modes we consider, we may do equilibration as well as
binding affinity estimation (to be discussed further at next meeting).
Optional - provide RC with more methodological details on the MD
benchmarks run to date using NAMD, if more details are available, prior
to the next meeting.
Current estimate of time required for MD simulations and computational
referee report revisions: 1.5 months, 50% effort (subject to change)
3) In addition to the above, I will also consider further: the minimum
additional work that is necessary to address reviewer concerns, effect
this work would have on progress on 2nd paper, how much of overhead work
would be synergistic with 2nd paper, how much of paper would need to be
rewritten. Assuming the relative binding affinities change after MD on
one set of structures, what would be implication for other reported
binding affinities?
4) We will have another meeting (PL and RC; optional for XG) regarding
details of required computational work (esp which simulations to
prioritize, duration of each simulation, and estimated computational
time required for these simulations).
We have up to 2 months to resubmit.
Raj
RC (11-20):
Software and hardware issues:
- GPUs: do we want to do serial or parallel computations?
Are there 16 cores on slave006?
We have multiple nodes. Assume you are referring to serial implementation on a single node/workstation. If we go with a in-house solution, we should consider either adding GPUs to an existing node or installing a new node which is GPU compatible. We should obtain a quote on a single prebuilt GPU compatible node (Wei Chen can obtain it once given the model information).
-What about amazon cloud ec2 gpu instances? ec2 also provides networked hpc instances for parallel computing. We may have use for these in other projects. Provide writeup on pros/cons of different approaches for RC to review.
Please provide estimate of required time for planned MD simulations above assuming single gpu.
PL to provide written info. RC can provide access to PMC-AT's current ec2 instances if needed.
- MD engine: NAMD or Desmond. Is corporate license required for latter? Friend wrote most of Desmond. Other friends use it for drug discovery w GPUs/cloud computing. Can inquire further about recommended configurations/speed improvements, including parallelization and some benchmarks. However, this is likely more relevant for lead optimization. Should we complete work for this paper with NAMD? Will there be any issues later switching to a new engine?
PL (11-14):
Hi, Raj,
Since we do intend to run MD simulations to compliment the experimental work, I think it is necessary we invest on some more powerful computing facility, especially the current GPU technology. Although we might not be using Amber for our simulation (the cost for license is too high), their website do provide valuable information on recommended hardware and benchmark information on using GPU vs CPU. http://ambermd.org/gpus/recommended_hardware.htm
There are vendors who make special GPU workstation or servers. With little budget, we can build our own system as suggested at the bottom of the page. I had experience before building computers from components, and we can quickly assemble one for the immediate needs.
For reference, they provided a benchmark results on simulation of DHRF (~23,000 atoms), and single GPU is 7 times more powerful than 16 CPU cores. For reference, we can barely obtain 1ns on our 16-core server for our system (45,000 atoms).
Let me know what you think,
Thanks,
Ping
PL (10/15): Raj, thanks for the updates. This week I will focus on the review of Sirtuin projects and prepare for the group meeting.
Here is what I plan to do next on the PCR OCT project.
1) Continue to review the codes (understand and confirm all the parameters);
2) Make necessary modifications to the code to reproduce some of the figures in BP and JCP drafts;
3) Communicate with Chaoran to get a better understanding what the experiment and theory.
Hopefully these tasks can be completed this month, and I will update from time to time.
RC (10-14): I have asked about some of the issues that we identified with the PCR codes. Answers below.
1) It seems the melting code has been written in such a way that a length-dependent k_1(T) is assigned based on the estimation that has been done for N=8,10 and 12 using experimental data. How was the expression provided for k_1(T) for all other lengths chosen?
Answer: kf is available till N = 14; for BP paper we can use N = 14 for primer length for illustrative purposes. For N > 14, the value for N=14 can be used as an approximation.
2) For sigma calculation for sequences where we do not have experimental data (i.e., for the sequences studied in the BP paper), are NN parameters to obtain Keq(T)?
Answer: Yes K is calculated based on NN parameters.
3) There appears to be only a two-sided melting code provided for DNA hybridization relaxation time calculations. I assume the part of this code that constructs the one-sided melting matrix was used for one-sided calculations, and did not prepare any separate code for one-sided calculations.
Answer: The one-sided melting matrix was constructed in a matlab function and the code was used in the construction of the two-sided melting matrix. PL: verify this and show you can use this matlab function for one-sided melting \tau.
4) Was the AT base pair stability constant used when computing the AU base pair stability constant for the sequences where experimental relaxation time data is available?
Answer: Apparently not. Craig et al data was used to calculate the AU stability constant. PL, please list the relevant data from this paper that can be used for AU stability constant calculation here. As I recall, the Craig et al paper did a steady state analysis, which included the use of the expression for the relaxation time in terms of the single stability constant s (since this is a homopolymer, there is only a single stability constant). It should be possible to solve for this stability constant.
5) In single_strand.m, line 26, assuming the single strands will recombine to form DNA, should the contribution should be min(y(1), y(length_template(1) + 5)), instead of min(y(1) + y(length_template(1) + 5)), which causes a sudden jump in [DNA] at the last cycle?
Answer: Yes this is a typo. Also, we are considering only N-1 cycles.
6) For enzyme binding: The temperature ranges in the code do not seem to match those in the latest BP paper figure that shows the \Delta H for two temperature ranges. Please explain the discrepancy in these temperature ranges.
Answer: When the Arrhenius relationship was fit, there were three different regions with different k0 and Ea values. In the \Delta H figure in the BP paper, two of these regions were combined for illustrative purposes. since they had the same slope.
7) What reference was used to obtain the value of kcatbyKn shown in the code?
Answer: kcatbyKn was estimated based on the nucleotide addition/second data in innis et al. Not sure exactly how this was done. See the extension section of BP paper for further info and plots.
8) From what reference did you obtain the value of kcat_r in the code?
Answer: Mehra et al
RC (10/13/13): I have posted meeting minutes and tasks below. In the future, please make sure to do this yourself.
Feel free to make changes or ask questions. RC (10/14): Please post your planned schedule for these tasks.
PL (10/4/13):
Some Docking results:
| Receptor |
Ligand |
Docking method |
Scores (MM-GBSA) |
Note |
| 4FVT-receptor (with pep) |
NAD+ ligs |
Glide SP 12 (top 2 rank) |
-105.4 |
|
| 4FVT-receptor (with pep) |
NCA-ligand |
Glide SP 14 (top 3) |
-38.1 |
Although +1 charged will have better binding affinity |
| 4FVT-NCA-1 (with pep, NAM) |
NAD+ ligs |
Glide SP 15 |
-81.4 |
NAD+ take a position somewhat close to B pocket but not. NAD (neutral NAM moiety) have similar binding affinity (-82.4) but NAM moiety take a different position. |
| 4FVT_enz-only |
NAD+ ligs |
Glide SP 16 (top 3) |
-104.6 |
NAD+ docked into the same position with similar binding affinity as with pep. |
| 4FVT-O-prep-5 (with pep, NAD+) |
NCA-ligand |
Glide SP 17 |
-28.5 |
NAM takes two slightly different position, the one closer to B pocket have a lower score at -24.1 |
| 3GLU-prep-3a |
Lys-pep |
Glide SP 10 |
-69.0 |
Rebind the short peptide back into the active site. MM-GBSA recapture the best rank |
| 3GLU-prep-3a |
Lys-pep |
Glide XP |
-69.6 |
Rebind the short peptide back into the active site. MM-GBSA recapture the best rank |
| 3GLU-prep-3a |
Lys-ac-4fvt |
Glide SP 18, 20 |
No near native pose found, (try using softer vdw radius in the future?) |
|
| 3GLU-prep-3a |
Lys-ac-4fvt |
Glide XP 19 |
No near native pose found |
|
| 4FVT_enz-only |
Lys-ac-4fvt |
Glide SP 21 * |
-102.9 |
Redock Acetyl-Lysine peptide back to 4FVT, peptide shift in position, not native found |
| 4FVT_enz-only |
Lys-ac-4fvt |
Glide SP 22 #5 |
-106.1 |
Found native structure after some slight change in docking protocol |
| 4FVT-O-NAD+ (with NAD+) |
Lys-ac-4fvt |
Glide SP 23 #7 |
-115.2 |
Found native structure |
| 3GLS-A-prep-4 |
NAD+ ligs |
Glide SP 24 * |
-84.4 |
No native structure found |
| 3GLS-A-prep-4 |
NAD+ ligs |
Glide XP 25 #3 |
-67.7 |
Near Native found |
| 3GLS-A-prep-4 |
Lys-ac-4fvt |
Glide XP 26 (SP 27) * |
No native structure found |
|
| 3GLS-NAD (with NAD+) |
Lys-ac-4fvt |
Glide SP 28 |
-68.4 |
Somewhat close to native structure found |
| 3GLS-NAD (with NAD+) |
Lys-ac-4fvt |
Glide XP 29 * |
-64.2 |
No native structure found |
| 3GLS |
NAD+ ligs |
Induced fit 02 * |
-97.1 |
No native structure found** |
| 3GLS-A-prep-4 |
NCA-ligand |
Glide XP 32 |
-29.9 |
Binds to C pocket |
- receptors are enzyme alone and/or with peptide substrate, and/or with NAD+, and/or with inhibitor (NAM)
- substrates include peptide ( Lys-pep), acetyl-lysine peptide (Lys-ac-4fvt) , NAD+, and NAM (NCA-ligand)
Keep in mind that since some scores are evaluated using the complex structure, the interactions are better optimized than using apo-enzyme or enzyme with other substrate.
What these results may suggest still require further discussion.
RC: Some of the results above are difficult to interpret. Please also provide more info on whether you are planning to use any scripting in order to customize the docking and binding affinity estimate calculations. It appears that even if full MD is not used, given the poor performance of some of the existing methods, some modified methods for sampling of conformational space of sirtuins (beyond just standard induced fit) may be required.
PL (10/09): My plan is to use Glide XP+MM-GBSA to select certain hits, then carry out MD simulations to obtain more accurate structural information and sampling, followed by MM/PB(GB)SA or LRM calculation for binding affinity. Currently, preparation is underway for MD simulations. I have installed AmberTool13, which includes codes for running MM/PB(GB)SA calculation, and NAMD, which is able to run MD. I will be doing some tests and see the performance first. If necessary, we can build a GPU workstation to get the computation power needed.
RC: Please plan to review the details of the methods, including the MD approach, at the group meeting. The workflow described by XG (as discussed with her) was not sufficiently descriptive in terms of how experiment would be used in conjunction with computation, and how experiment and computation would both be applied in the immediate follow up paper, for which we already developed a plan that we would like to follow through on. This also needs further explanation. Also, regarding using straight docking for hits, given the inability of this method to often successfully reproduce xtal data above, it is not clear whether using docked structures directly in MD without some additional faster conformational sampling of the type described above is sufficient - please address this as well.
PL (10/11): Will work on presentation for group meeting and will include these topics.
1. Sirtuins Project: (Proposed research and methods.)
1a. Applying Docking Method and MM-GBSA in analyzing binding modes and binding affinities for various substrates in SIRT3.
SIRT3 has several crystal structures available now. By comparing various crystal structures, we noticed that there are significant structural differences when different substrates are bound, and the loop and helix (residues 155-169) are the region change most. They can be classified into three categories based on the bound substrates.
A. 3GLS (SIRT3 strcuture) 3GLU (SIRT3 with AceCS2 Peptide), 3GLR (SIRT3 with acetyl-lysine AceCS2 peptide), 4FZ3 (SIRT3 with acetyl-lysine P53 and fluorophore attached),
B. 3GLT (SIRT3 with intermediate analog), 4BVE (SIRT3 with intermediate analog), 4BVF (SIRT3 with intermediate analog), 4BVG (SIRT3 with intermediate), 4BV3 (SIRT3 with ex-527 and NAD+),4BVH (SIRT3 with AADPR and EX-527) 4BN4 (SIRT3 with ADPR),
C. 4FVT (SIRT3 with NAD+ analog and Ac-ACS peptide), 4JSR (SIRT3 with inhibitor), 4JT8 (SIRT3 with inhibitor), 4JT9) SIRT3 with inhibitor), 4BN5 (SIRT3 with SRT1720 Inhibitor)
To completely understand the inhibition mechanism, it is necessary to understand various binding modes and binding affinities under different conditions, i.e. with or without Acetyl-Lysine Peptide, with or without NAD+, with or without inhibitors. Enzyme-Substrate-Cofactor-Inhibitor complex needs to be completely analyzed. While certain starting crystal structures will fail for the formation of certain complex, other structures may works out. Recent publication on Journal of Chemical Information and (attached.) compare different docking protocols and multiple PDB receptors using various receptors structures may improve the docking performance. ci400040d.pdf
Docking and MM-GBSA calculations using various starting structures is currently underway.
RC (9-26): Please provide some more details on the methods you will use to accurately estimate binding affinities. As you have seen in this paper, MM-GBSA without MD is not capable of predicting absolute binding affinities without a training set. Are you planning to run MM-GBSA calculations without MD and with limited sampling, as Eric began to do?
Q: Are you planning to run regressions on congeneric series of ligands for the purpose of prediction; and if so, which/how many?
PL (10/02): Not right now. The use of congeneric series of ligands for prediction works under the condition that the receptor is positioned ready for docking and assuming difference between the congeneric series reflect the difference they will have in correct binding. Currently, this condition is not confirmed. And the ligands of interest do not fit into this category.
RC: If we are interested in accurate binding affinity prediction, the alternative would appear to be full MD for each ligand. Since the plans for connecting experment with theory in this paper hinge on the approach used, we should prioritize the determination of whether prediction methods of this type are viable for the small molecules we are planning to look at in the lab. It would seem that at least some of the small molecules that bind in the C pocket should be congeneric. Please specify why the ligands of interest do not fit into this category.
PL (10/02): My plan on this project is first to further investigate the inhibition mechanism, specific to different type of inhibitors. Therefore we can explore all the possibilities in small molecule drug design and correlate the experimental observation with the modeling/simulation. For example, Ex-527 inhibits Sirtuins after intermediate formation, but the exact mechanism remain to be answered, and previous docking study suggest Salermide-type inhibitors bind to the same binding pocket of ribose and nicotinamide moieties of NAD+. And these are two molecules Xiangying has studied experimentally. Because inhibition works in different ways, we need to investigate the inhibitor binding under different conditions, i.e. with or without NAD+/Acetyl-lysine peptide/intermediate/products. In term of inhibition mechanism and potency, the small molecules provides little values, because, 1) the size of these molecules are too small compared to the binding pockets, and without acetyl-lysine peptide and/or NAD+ bound, they can bind in many locations, 2) they usually bind with relative low binding affinity except NAM. As we can see from previous LIE and MM-GBSA studies, as well as our own docking studies, the accuracy of binding affinity is hindered in large part by the inaccurate protein structure upon binding, that is the reason we need MD to take into account induced protein structural change and dynamics sampling. Also, neglecting entropy contribution is probably the cause for large differences between current MM-GBSA results and experimental values, which may be estimated using normal-mode analysis. Entropy contribution will be significant when trying to produce more accurate binding free energy and comparing molecules with different size and binding modes.
RC (10-2): Since the plan for the project may be changing compared to our previous plans, please have a meeting with XG soon and afterwards post how the new plan is related to the old plan that we have discussed. We need to ensure that the experimental plans are aligned with the computational ones. In particular, if MD is required and there is no experimental parameterization of model parameters to be done, it is not clear that computational screening is faster than experimental screening. Thus these changes may affect overall plans for the project.
Regarding small molecules, we had some interest in predicting binding affinities of these as leads for activators. Please discuss this with XG as well.
Q: In this regard, are you planning to run multiple linear regression (LRM without MD)?
PL (10/02): There is no point in rushing into LRM if the applicability is not confirmed.
Q: What about induced fit - what type will be used? Exactly how will it fit into your protocol?
PL (10/02): I have run some tests with induced fit, with different setting, in trying to test its performance on NAD+ docking to free SIRT3 crystal structure or crystal structures with different substrate. The results indicate that for flexible ligand of the size of NAD+, induced fit is not able to produce correct pose. Instead, simple Glide using SP or XP setting can capture the correct poses that rank near the top on Glide score and rank top on MM-GBSA function. The plan on improving the docking results is using MD for complex structure optimization.
RC: What about for smaller ligands of the size of NAM?
PL (10/02): For free SIRT3, there are many places NAM can bind to, and the C-pocket is already positioned to the favorable place for NAM binding. Induced fit do little to improve the results, and will likely populate more on the false positives.
Q: How will you ensure the results will be comparable across ligands? What will you define as a congeneric series - same binding mode? Hence, Ex-527 would not fit into the same series as NAM...
PL (10/02): To ensure compatible results for various ligands, we have to make sure correct pose is captured in the docking and further refinement of the structure using techniques such as MD to ensure all important energy contribution are included. Congeneric series share same binding mode, and often share some functional groups, and often have similar poses.
PL (10/02): So far, I have completed some docking calculations using 4FVT as starting receptor structure, and generate single substrate structures (NAD+, NAM, Acetyl-Lysine Peptide) and complex structures up to all three substrates. The binding modes and corresponding MM-GBSA suggested some interesting finding. (Will summarize soon.) Docking using 3GLS (free SIRT3) and 3GLU (SIRT with Peptide) show different performance, and 3GLS based docking are able to capture correct poses for NAD+, but requires further structural refinement to improve the MM-GBSA function. (Induced fit doesn't work as expected.) However, binding of Acetyl-Lysine Peptide in 3GLS requires further work. After further investigation, I plan to set up the cluster for MD simulation. After checking the license information for various software and capabilities, I plan to use ambertools to prepare the simulation and NAMD to run the simulation. Currently, there is no MPI implementation on the cluster, which I plan to add on to improve the performance.
Will you run MM-GBSA on congeneric series without MD, check the predictive power, and then run MD if the correlation is low?
PL (09/27): Yes.
When do you think it would be feasible to start writing an outline of a methods section (if only for internal use as a progress report)?
PL (09/27): In less than two weeks.
PL (09/26/2013): After inspecting some docking results using 3GLS and 4FVT, it seems running some MD simulation maybe necessary to improve the results. With 4FVT (SIRT3 with NAD+ analog and Ac-ACS peptide), it is easier to redock NAD+, with or without Ac-ACS peptide, and the MM-GBSA results are similar, and it can also dock NAM into C pocket with Ac-ACS peptide and much lower MM-GBSA binding energy, and NAD+ can also be docked into A pocket and nicotinamide motif can fit into other position than B or C, with somewhat decreased MM-GBSA binding energy. It would be interesting to further explore various binding mode with substrate/co-factor/inhibitor in different places. Using Glide on 3GLS can also produce reasonable structural pose for NAD+, but not difficult for Ac-ACS peptide, and the MM-GBSA binding energy is weaker as well, mainly due to the failure for SIRT3 to adjust upon binding. Therefore, it is desirable to start with a crystal structure pre-positioned to accommodate the ligand and a method for improving the result on both structure and energy. (Details to be summarized after a whole series is completed.)
Using single structure from docking (Glide or induced fit) works only for some specific cases, and may not be able to generalize to new ligands, which may induce larger receptor changes after obtained from docking. The prediction on either structure or energy/binding affinity is relative large uncertainly simply from docking. One possible MD code to use is NAMD, which is capable of using implicit solvent model such as GB for speedup. Desmond is more straight forward, but it uses only explicit solvent. Without correct prediction on structure, LRM results will carry a large uncertainty as well. It is too early to run any calculation on ligands if the docking result can not produce the desired accuracy.
1b. A good selection of substrates and binding affinities are required for LRM method and will be good for more accurate binding affinity estimation of congenic series. (A future direction, not urgent for mechanism study.)
2. PCR Project:
2a. Using the existing code to reproduce BP figures.
Progress: Using same settings, and with slight modification to the code, I am able to generate a figure similar to Figure 8 in the paper.
Need to find out more details about the setting. Modification of the code to reproduce other Figures are underway.
RC (9-26): Thanks. Please post another update today, regarding the unresolved issues regarding required settings (i.e., what additional info you need that is missing in the transferred files), a more detailed analysis of what you are plotting in each case and why, and what your plan is with these codes for the next few days.
PL (9-26): There is melting process included in the current code, which seems to affect the Enzyme concentration only (by enzyme deactivation). And with 30 second each on annealing and extension, I didn't get the same efficiency as plotted in Figure 8. (Maybe the reaction parameter changes?) With current set of reaction parameters, better efficiency is achieved with longer annealing time and shorter extension time (i.e 55:5).
RC: Is this the only test you have run this week? Can you tell me how much time you are spending per day.
PL: This is the one I think is with correct results. There is the other codes I modified showing inconsistent results. I spent average two hours on PCR project, four hours on Sirtuin project, two hours in literature search.
PL (09/27): In Figure 9, for Staged Time Invariant (ON-OFF model) PCR (Mehra and Hu), DNA won't form in the annealing step (the first 30 minutes), isn't that right? But the figure shows otherwise. On Figure 9 to 12 and 16, 19, with various setting, the system always achieves maximum DNA concentration, which seems somewhat counterintuitive. I didn't observe it in my simulation, am I missing something here?
The next step is modifying the state equations to model "Staged Time Invariant, Time Invariant, and Time variant PCR model).
RC (10-2): A list of additional questions is being prepared by PL based on review of codes vis-a-vis latest JCP draft.
PL (10/02): Currently there are some details need to be clarified on the PCR Optimal Control code. The documentation only states briefly what each function does, but not much about the mathematics background, which will take me longer to digest. Also there is some values used in the code I am not sure about the origin, i.e. the reaction parameters (where do the differences in reaction parameters for [S1] and [S2] come from?) , the lower boundary for temperature (u(i) has a lower boundary of 0.0233). And I am also checking out the simulation code for simplex PCR as well. I am reading general Optimal Control theory as well. If you have any book/reference to suggest, please let me know.
RC (10-2): Yes, I will provide the necessary background reading. Before that, it should be possible to run many tests on the parameter estimation and model simulation parts of the work to date. The JCP and BP papers do not apply any optimal control algorithms to generate any of the results. Thus we would like to see a list of questions regarding these aspects of the work and data reproduction for JCP and BP that can be communicated to Karthik. Ability to reproduce these results will be important for further work on optimal control. Thus please prioritize this before getting involved much further in OCT theory.
In general, a schedule of work will be useful.
PL (10/02): I believed I have a good understanding of the kinetic model used in PCR OCT code Karthik originally sent, and I am able to modify the code in order to reproduce some of the figures in the drafts. However, the parameters in the code are probably not the ones used in the draft, which make it difficult to reproduce those figures exactly.
In this week and next week, I will try to finish digesting both codes and understand the flow of the codes as to the kinetics model applied, and run tests on the task you suggested (parameter estimation and model simulation). And I am also reading other papers on kinetics model and on optimal control theory.
RC (10-2): As the priority, I would like to see a daily (or every 2 days) schedule for testing that will be done with these codes, which parts of which paper each of the tests is related to, and a list of unresolved questions pertaining to each (such as the ones you have listed above). It is ok if putting together this plan takes a day or so. Regarding OCT, I have taught classes on this and have detailed lecture notes that can be provided. I suggest you let me know when you are reading about OCT so I can provide this. Regarding other literature, in general, please indicate what aspects you are looking at so I can determine whether I should provide direction in this regard based on the past work we have done.
In general, please plan to provide a detailed schedule and updates on the wiki at the frequency of every 2 days or so, so I can keep abreast of and help direct the work where needed (we may find that this high frequency is ultimately unnecessary once we get a clearer picture of where each project is headed). Thanks.
RC: Please post this schedule with planned milestones/questions and answers to other 10-2 questions first thing on Mon.
PL (10/07): The schedule for PCR OCT project:
The two codes I received from Karthik covers different aspects related to the PCR OCT project, and are related in part to the BP and JCP draft to some extend. However, not all pictures are generated using these two codes, and further inputs (i.e. model and parameters) or modifications of the codes may be required. The current plan is based on the two codes provided.
Simplex simulation Code:
1) Understand the two-side melting model for the generation of parameters of sequence-dependent oligopeptide hybridization kinetics. (rate_constants_annealing_2s.m and associated functions, including the examination of oligoprop.m code I wrote.) ~ 3 days.
Question 1: in evolution_species_prc.m, line 38, fraction_time = time_exten/reaction_time; shouldn't it be fraction_time = time_anneal/reaction_time; as it was used to determine which rate_constants to use in state_equations_pcr.
Question 2: in single_strand.m, line 26, assuming the single strands will recombine to form DNA, the contribution should be min(y(1), y(length_template(1) + 5)) instead of min(y(1) + y(length_template(1) + 5)), which cause a sudden jump in [DNA] at the last cycle.
2) Using the simplex code to simulation PCR kinetics under various conditions, ( temperature, time, sequence, PS/ratio, etc. Experimental results, if any, will help greatly in validating the model) ~ 3 days.
Question. Experimental validation in Figure 31 (JCP draft) is hard to read and the steady-state model performs well in Figure 31, while in Figure 5 to Figure 11, there are significant difference between experimental and steady-state model. Details for generating these plots may help.
3) There are models/simulations , i.e. in case of mismatching occurs, are not in the code. If not provided, it will take extra time for coding. ~ time undecided.
RC: The first 9 pages of the JCP draft should be the focus of reproduction efforts. The rest is still under revision and some of it is obsolete. I would like to see a list of specific paper figure reproduction efforts underway with the results and discrepancies posted to the wiki periodically (every 2 days, e.g.). I can ask for the missing codes for specific figures if you can make a specific list of which figures.
PL (10/09): Again, the simulation code is used to simulate the PCR cycles, it is able to generate rate constants and some relevant parameters from preset parameters.
The code used to estimate of model parameters was not provided.
RC: Yes, I understand the role of the simulation code as you have mentioned. I thought you received another code that was used for the JCP parameter estimation. But it seems he only provided the code for computing the relaxation times/rate constants from the two-sided model. Please confirm. In that case I will seek to get the model parameter estimation code. It has been written.
PL (10/11). Yes. The code provided only includes computing relaxation times/rate constants from two-sided model only.
RC: Please list all the parameters and associated rate constants here. Regarding enzyme binding as well as DNA hybridization, I would like to see the details of what is in the code and what is missing.
PL (10/11): The current parameters used to determine rate constants:
NN parameters from this paper, "SantaLucia Jr., J. (1998). A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proceedings of the National Academy of Science USA 95, 1460–1465.", used to determine the free energy and equilibrium constant for DNA hybridization, and equilibrium concentrations for [S] and [P].
The so-called forward_rate_constant is given by the formula below, not sure where it comes from:
if N == 8
kf = exp(6671.2*(1/(T+273)) - 2.6229);
elseif N == 10
kf = exp(7511.4*(1/(T+273)) - 5.2212);
elseif N == 12
kf = exp(8179.1*(1/(T+273)) - 6.7862);
else
kf = exp(11498*(1/(T+273)) - 17.602);
end
--------------
stability constant and sigma calculated using parameters from {J. G. Wetmur and N. Davidson, Journal of molecular biology 31,349 (1968).} which apply to A-T and G-C types of base pair, and the equilibrium constant obtained above.
relaxation_time is determined using all the information obtained above.
Then the overall forward and reverse rate constants for DNA hybridization are determined.
RC: Was there code provided for calculation of sigma using the method above?
PL (10/14): Sigma is simply K_eq/prod(stability_constants). It is coded.
RC (10-11): I believe what is called kf in the code is actually k_1 in the paper (the notation was changed later). This is the forward rate constant for an individual base hybridization step. The important point to note is that k_1(T) is being --estimated-- and then the estimates used for prediction of relaxation times for sequences of the same length in several of the figures in the JCP draft. It appears that in the code sent above, fixed values for kf are used for particular sequence lengths N, based on an Arrhenius relationship to model the temperature dependence. This Arrhenius relationship was shown in several of the figures in the JCP paper.
The estimation code was not sent. In summary, it solved for k_1(T) (kf(T) in notation above) given the tau(T), sigma(T), and stability constants, together with the one/two-sided/steady-state model, via nonlinear root finding. The assumption was made that k_1(T) does not vary with sequence, only length. Thus is was possible to use the same k_1(T) estimated on the basis of one sequence to predict the relaxation time for another sequence of the same length.
For enzyme binding: The current code and parameters are used: (not sure where the parameters come from)
if T >= 5 && T <= 30
Ea_forward = 3807.5;
k0_forward = exp(29.66);
Ea_reverse = -584.73;
k0_reverse = exp(-3.7603);
elseif T>30 && T <= 50
Ea_forward = -2490.5;
k0_forward = exp(8.8811);
Ea_reverse = 484.57;
k0_reverse = exp(-0.2393);
else
Ea_forward = -11592;
k0_forward = exp(-19.198);
Ea_reverse = 735.12;
k0_reverse = exp(0.547);
end
RC (10-11): In the BP paper, you should note that there is a section on modeling of the enzyme binding rate constants based on certain assumptions including the activation energy and enthalpy of reaction being of the same order of magnitude (I hope you have the most recent version that includes this). As you can see from that section, the forward and reverse rate constants for enzyme binding were only known at one temperature. It was necessary to estimate the rate constants at other temperatures based on some model. As I recall an Arrhenius model was used (preexponential factors and activation energies are given above). A given Arrhenius model (given k0,Ea) only applied in a given temperature range (this should be apparent from the variation of delta H_rxn with temperature shown in a figure in that paper, along with the above assumption regarding the reln between Ea and delta H_rxn).
The delta H_rxn came from other experimental literature. (Meeting task: You can list the ref here when you find it; should be cited in BP).
I assume the simulation code computes forward and reverse rate constants for binding based on the above Arrhenius parameters.
RC: Meeting task - Please check that section of BP to make sure that all steps of the forward and reverse binding rate constant calculation methodology described in that section are actually included in the code. (Were there any equations in that section that included the relaxation time for enzyme binding?)
Meeting task - look at (or discuss w Chaoran, who read it) the Datta/Licata paper that has data on the forward and reverse reaction rates for enzyme binding at a particular temperature.
For extension, the rate constant is determined this way. (Described in BP draft, but didn't mention what system it applies to)
kcatbyKn = exp(48.29)*exp(-12268/(T + 273));
kcat_r = 10; (This one is for enzyme/DNA dissociation. But can't find any reference for this constant.)
RC (10-11): This is fine. The kcatbyKn (from Michaelis-Menten theory) was an approximation based on data available in the literature, but is not an accurate value. Chaoran is now doing new experiments to obtain kcatbyKn(T) more accurately.
Regarding kcat_r, I also don't recall where it came from. How does it compare to the reverse rate constant for enzyme binding to D_i (partially extended primer-template) above?
RC: Meeting task: Discuss with Chaoran her work on determining kcatbyKn(T) so you can later use it in the code.
In JCP draft, figure 3-11 are for oligoribonucleotides of form AnUn, and simulation code only deal with A-T, G-C pairs, therefore the parameters in the simulation code don't apply.
RC: The parameters (e.g., sigma, k_1) used in predicting relaxation times for DNA hybridization were supposed to be length dependent but not strictly sequence dependent. Are these the parameters you mean? Please be specific here. The sequence dependence of sigma can come from equilibrium thermodynamic stability of the DNA sequences, which can be computed. Please also explain what is meant by "simulation code only deals with A-T, G-C pairs". The experimental data on AnUn was, I believe, used to estimate kf for sequences of such lengths, and to test the model predictions of relaxation times for other sequences of the same lengths.
RC: I will also be sending you an updated draft soon.
PL (10/11): The stability constants and equilibrium constant are sequence dependent, (see the listed code above), and sigma and k_1 are sequence dependent as well. And no parameters is available for "U" in the simulation code.
RC (10/11): As noted above, k_1 is intended to be length but not sequence dependent; however, there is an issue with the way k_1 estimation was implemented in the paper, which I have raised as a point for ongoing revisions - namely, it was estimated based on just one sequence, and subsequently assumed to be the same for other sequences (in this sense, it depends on the arbitrary sequence that was used to estimate it).
Regarding parameters for "U" in the simulation code, we may need to discuss what you mean. To compute the forward and backward rate constants for annealing for a given sequence, you need the relaxation time, which depends on sigma, k_1, stability constants. k_1 as mentioned should be length but not sequence dependent. According to the code above, it appears k_1 is assumed to have the same temperature dependence for any sequence length above a certain length. sigma is currently assumed to be sequence dependent through the sequence dependence of overall stability constant and Keq (the latter could be computed based on NN parameters for the time being). If these parameters are not available for U, have you tried to assume the values are the same as those for T? Please explain if you mean other parameters. Also, why do we want the parameters for U, since in the BP paper, we are generally interested in sequences that do not contain U? Thus, we would want sigma for these sequences and k_1 for any sequence of that length.
RC: Meeting task - use the stability constant for AT base pairs in place of AU and try to match the results in the paper.
Meeting task - see if you can find the experimental K_eq's for these AU sequences from the paper cited.
Also, the simulation code use only two-side melting mechanism, codes for one-side melting or steady state are not provided.
RC: I can try to get the one-sided and steady state codes.
RC: Update based on meeting - the one-sided code is part of the two-sided code. Meeting task: learn how to apply the one-sided model using the code provided.
Figure 3 was not referred in the text, and not sure where the data comes from. Data in Figure 4 - 11 also require some other relevant parameters, which are not mentioned in the draft. If the current target is to reproduce the figure, I will need relevant codes.
RC: Please list the parameters - do you mean sigma and k_1? See above.
If the goal is to understand the code provided, it is not very practical to have updates every few days, because, from time to time, I need to dig into the literature to find out where the origin of the data or the formula is from, and they are not always supplied in the two drafts I had. It is probably more practical to set a goal, i.e. for the simulation of PCR cycles, understand the code, being able to clean it up and make it more readable, being able to make modifications according to the change of kinetics model.
RC: Rather than read up on literature independently, as indicated, you should inquire with me first on where the formulas or data are from. If you do not need to search the literature independently and we communicate according on specific points as above, this should make it possible to have frequent updates of the type requested.
RC: Please list your updates using bullet points rather than paragraph text to make clear the outstanding issues each day (now separated above).
With the simulation code, I can generate sigma vs 1/T for A_7T_7 and G_7C_7, but it is not the one in Figure 3.
PCR OCT code:
1) Understand the kinetic model (state equations) used in the OCT code, modification of the code for different models (staged, time-invariant etc, reaction parameters required for reproducing the figures). ~ 3 day.
2) Derive corresponding costate equations, control constaint, objective function, and understand the optimization routine. ~ undecided.
3) Modified the code for other conditions. ~ undecided.
Time is also needed to learn about background of optimal control theory.
RC: As noted above, 2,3 should not be pursued at this time, and I will provide necessary guidance regarding materials to use at the appropriate time.
References on experimental and previous theoretical studies on PCR related kinetics are collected and studies.
RC: As noted above, please list the references here.
RC (10-10): Not yet addressed.
PL (10/11): Currently I am reading all the references provided in the JCP and BP draft.
Time is also needed to learn about background of optimal control theory.
RC: As noted above, this should not be pursued at this time, and I will provide necessary guidance regarding materials to use at the appropriate time.
A good understanding of PCR experiments is needed as well for solving the real problems we are facing.
RC: We have this understanding.
Note: the current JCP draft is not completed and somewhat hard to read.
2b. More codes are needed.
RC (9-26): Agreed. I will follow up.
RC (10-2): The codes are now available.
PL (10/03): In the simplex PCR simulation code, one of the key function oligoprop is from MATLAB's Bioinformatics Toolbox, which we currently do not have. The cost for the toolbox is $1000. The toolbox is used in the code to get the free energy for DNA hybridization using NN parameters from this paper, "SantaLucia Jr., J. (1998). A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proceedings of the National Academy of Science USA 95, 1460–1465.", I can probably implement it myself, but it may take at least two days to get all the information and code ready. Also. if SaltValue is provided, oligoprop should return entropy after salt concentration correction, and the extra term in function eq_const may be redundant and overestimate of the entropy change.
RC: Will see if we can buy this.
PL (10/09): My own code is working fine so far. It is not required for now.
RC: Without further details on your implementation, I can't verify that the implementation will not introduce sources of error in our work. Please provide some details that indicate that the implementation will produce results essentially identical to those in the toolbox. Does it address any/all types of structural motifs supported by the toolbox?
PL (10/11): Here is the code I wrote:
----------------
function thermos = oligoprop( sequence )
% oligoprop to replace the original function in MATLAB's Bioinformatics Toolbox
% oligoprop returns thermodynamics information for DNA hybridization,
% delta_H and delta_S
% AA/TT (TT/AA) AT/TA TA/AT CA/GT TG/AC GT/CA AC/TG CT/GA AG/TC GA/CT TC/AG CG/GC GC/CG GG/CC (CC/GG)
keySet_NN = {'AA', 'TT', 'AT', 'TA', 'CA', 'TG', 'GT', 'AC', 'CT', 'AG', 'GA', 'TC', 'CG', 'GC', 'GG', 'CC'};
Hvalue_NN = [-7.9, -7.9, -7.2, -7.2, -8.5, -8.5, -8.4, -8.4, -7.8, -7.8, -8.2, -8.2, -10.6, -9.8, -8.0, -8.0];
Svalue_NN = [-22.2, -22.2, -20.4, -21.3, -22.7, -22.7, -22.4, -22.4, -21.0, -21.0, -22.2, -22.2, -27.2, -24.4, -19.9, -19.9];
dH_NN = containers.Map(keySet_NN, Hvalue_NN);
dS_NN = containers.Map(keySet_NN, Svalue_NN);
% G-C A-T
keySet_ini = {'G', 'C', 'A', 'T'};
Hvalue_ini = [0.1, 0.1, 2.3, 2.3];
Svalue_ini = [-2.8, -2.8, 4.1, 4.1];
N_in = ['G', 'C', 'A', 'T'];
N_inv = ['C', 'G', 'T', 'A'];
dH_ini = containers.Map(keySet_ini, Hvalue_ini);
dS_ini = containers.Map(keySet_ini, Svalue_ini);
% symmetry correction
dS_sym = -1.4;
seqin = upper(sequence);
seqlen = length(sequence);
delta_H = dH_ini(seqin(1));
delta_S = dS_ini(seqin(1));
for index = 1:seqlen-1
% disp(seqin(index:index+1));
delta_H = delta_H + dH_NN(seqin(index:index+1));
delta_S = delta_S + dS_NN(seqin(index:index+1));
end
seqin_r = '';
for index = 1:seqlen
N_index = find(N_in, seqin(index));
seqin_r = strcat(seqin_r,N_inv(N_index));
end
if strcmp(seqin,seqin_r)
delta_S = delta_S + dS_sym;
end
thermos = [delta_H, delta_S];
If the reference is right, I believed my code should produce the same results as in the toolbox. I am not against buying the toolbox, especially if there is anything you want to use in the toolbox. and it can certainly be used for testing my code.
RC: It would be best if you could compare the \Delta G's computed for a given sequence with your code to those reported in the literature or in our current papers that used the toolbox. There might also be some tools online for computing \Delta G's for given sequences using NN parameters. You can compare with those as well. Also, what about SaltValue mentioned above?
PL: 09/13/2013:
1. Analyze the interaction in new SIRT3/Ex-527 structure.
2. Analyze the inhibitor library Xiangying has and pick the most representative molecules for experiments.
PL: 09/09/2013:
1. Help complete the NSF report.
PL: 09/05/2013:
1. Prepare for docking/binding studies on small inhibitors XG is studying using Induced Fit Docking/MM-GBSA/LRM methods on SIRT3.
PL: 09/10/2013: Testing Glide and Induced Fit on 1YC2:A (In progress, 09/12/2013)
PL: 09/03/2013: Immediate task
1. Reviewing the "Mechanism of Inhibition of the Human Sirtuin Deacetylase SIRT3: Computational and Experimental Studies" paper by Xiangying Guan et al. (09/05/2013, First round of comment sent.)
2. Learn about the Optimal Control of PCR by reading "Sequence-dependent modeling of DNA ampli cation kinetics: toward optimally controlled polymerase chain reactions" by Karthikeyan et al
Learn about the associated code and run some simulations to test and understand the code. (PL: 09/09/13. Code obtained, in the process of learning. 09/10/2013, run the Code in MATLAB)
Set up meeting with Karthikeyan to discuss about the project.
3. Get license for schrodinger suite of program and get it up and running (DONE as of 09/04/2013, an updated version of Schrodinger Suite is also installed on my Desktop using remote license server)
4 Get a quote for matlab license and get it installed and up and running (MATLAB obtained and installed on Desktop PC on 09/10/2013, a linux version will be installed soon)